Liana Merk
@bacteriyay.bsky.social
99 followers
120 following
9 posts
PhD student in Sean Eddy’s lab! I like introns, phage, fermentation, and stamps.
Posts
Media
Videos
Starter Packs
Pinned
Liana Merk
@bacteriyay.bsky.social
· May 24

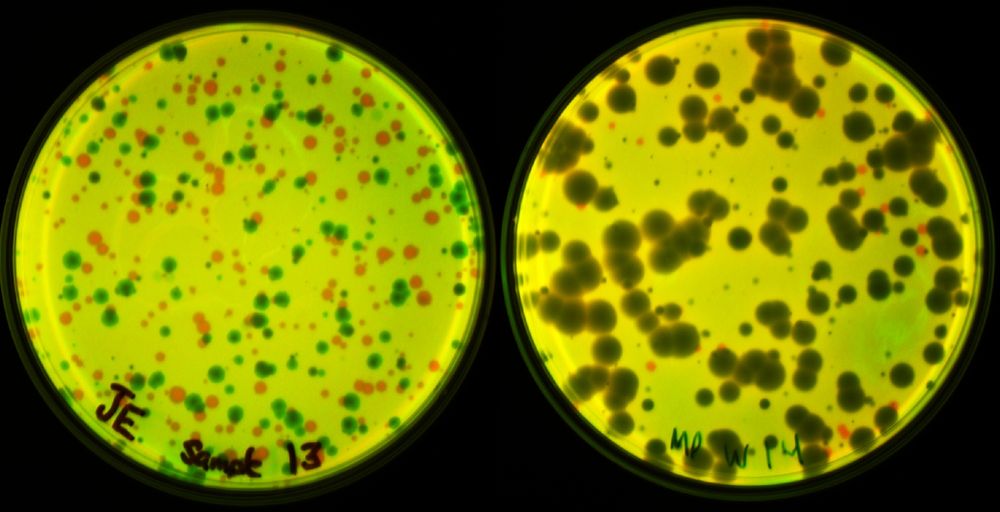
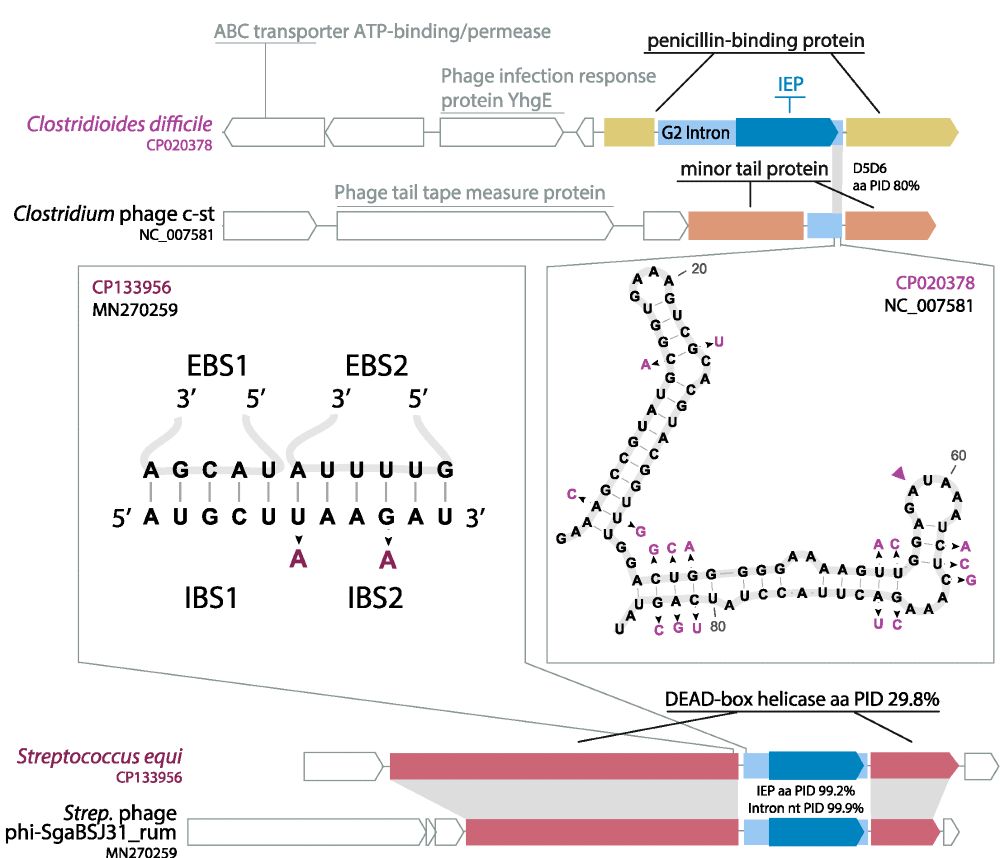
Prevalence of Group II Introns in Phage Genomes
Although bacteriophage genomes are under strong selective pressure for high coding density, they are still frequently invaded by mobile genetic elements (MGEs). Group II introns are MGEs that reduce h...
www.biorxiv.org
Reposted by Liana Merk
Reposted by Liana Merk
Michael Baym
@baym.lol
· Sep 1

Genomic resistance in historical clinical isolates increased in frequency and mobility after the age of antibiotics
Antibiotic resistance is frequently observed shortly after the clinical introduction of an antibiotic. Whether and how frequently that resistance occurred before the introduction is harder to determin...
www.microbiologyresearch.org
Reposted by Liana Merk
Reposted by Liana Merk
Sean Eddy
@cryptogenomicon.bsky.social
· May 28
Reposted by Liana Merk
Liana Merk
@bacteriyay.bsky.social
· May 24
Liana Merk
@bacteriyay.bsky.social
· May 24
Liana Merk
@bacteriyay.bsky.social
· May 24

Prevalence of Group II Introns in Phage Genomes
Although bacteriophage genomes are under strong selective pressure for high coding density, they are still frequently invaded by mobile genetic elements (MGEs). Group II introns are MGEs that reduce h...
www.biorxiv.org
Reposted by Liana Merk
Conner Langeberg
@clangeberg.bsky.social
· May 15

Tick-borne flavivirus exoribonuclease-resistant RNAs contain a double loop structure - Nature Communications
Many flaviviruses generate specialized non-coding RNAs that resist degradation by host exonucleases, promoting viral infection. Here, the authors characterize an RNA element in Powassan virus, highlig...
www.nature.com
Reposted by Liana Merk
Nick Polizzi
@nickpolizzi.bsky.social
· Apr 28

Zero-shot design of drug-binding proteins via neural selection-expansion
Computational design of molecular recognition remains challenging despite advances in deep learning. The design of proteins that bind to small molecules has been particularly difficult because it requ...
www.biorxiv.org
Liana Merk
@bacteriyay.bsky.social
· Apr 28
Reposted by Liana Merk
Reposted by Liana Merk
Reposted by Liana Merk
Fernando Rossine
@fernpizza.bsky.social
· Feb 21

Intracellular competition shapes plasmid population dynamics
Conflicts between levels of biological organization are central to evolution, from populations of multicellular organisms to selfish genetic elements in microbes. Plasmids are extrachromosomal, self-r...
www.biorxiv.org