Rui Yang
@ruiyang.bsky.social
71 followers
130 following
3 posts
Postdoc in Computational Biology @Buenrostro Lab | 3D Genome & Epigenomics | PhD @Leslie Lab
Posts
Media
Videos
Starter Packs
Rui Yang
@ruiyang.bsky.social
· May 15
Reposted by Rui Yang
Jian Zhou
@zhou-jian.bsky.social
· Feb 19
Reposted by Rui Yang
Reposted by Rui Yang
Reposted by Rui Yang
Danfeng Chen
@danfengc.bsky.social
· Jan 6

Coancestry superposed on admixed populations yields measures of relatedness at individual-level resolution
The admixture model is widely applied to estimate and interpret population structure among individuals. Here we consider a "standard admixture" model that assumes the admixed populations are unrelated...
www.biorxiv.org
Reposted by Rui Yang
Anshul Kundaje
@anshulkundaje.bsky.social
· Nov 21

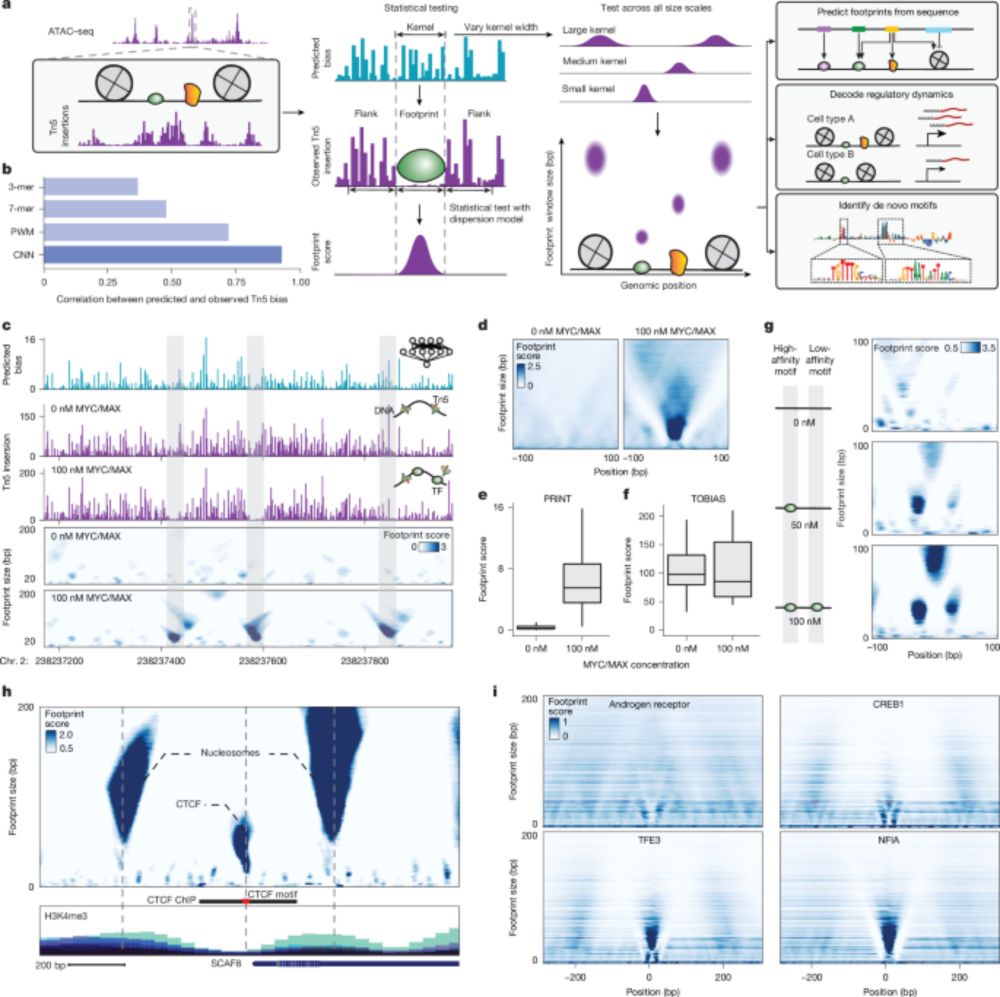
Single-molecule states link transcription factor binding to gene expression - Nature
A study uses single-molecule footprinting to measure protein occupancy at regulatory elements on individual molecules in human cells and describes how different properties of transcription factor bind...
www.nature.com
Reposted by Rui Yang
Mitch Guttman
@mitchguttman.bsky.social
· Nov 27
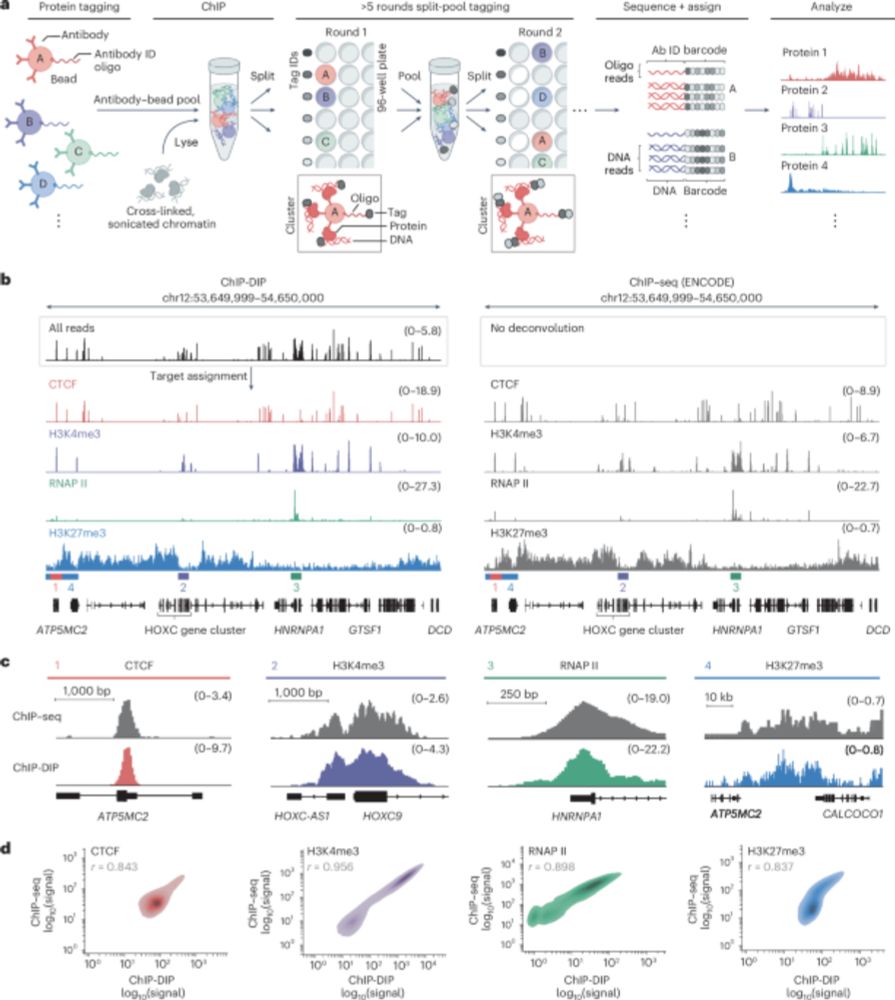
ChIP-DIP maps binding of hundreds of proteins to DNA simultaneously and identifies diverse gene regulatory elements - Nature Genetics
ChIP-DIP (ChIP done in parallel) is a highly multiplex assay for protein–DNA binding, scalable to hundreds of proteins including modified histones, chromatin regulators and transcription factors, offe...
www.nature.com
Rui Yang
@ruiyang.bsky.social
· Nov 26
Rui Yang
@ruiyang.bsky.social
· Nov 26

HiC2Self: self-supervised denoising for bulk and single-cell Hi-C contact maps
Hi-C is a chromosome conformation capture assay used to study 3D genome organization. The recent development of single-cell Hi-C technologies has further enabled the examination of 3D chromatin organi...
www.biorxiv.org
Reposted by Rui Yang
Su-In Lee
@suinlee.bsky.social
· Nov 23
Reposted by Rui Yang