
go.bsky.app/QVPoZXp


rdcu.be/eFnIc
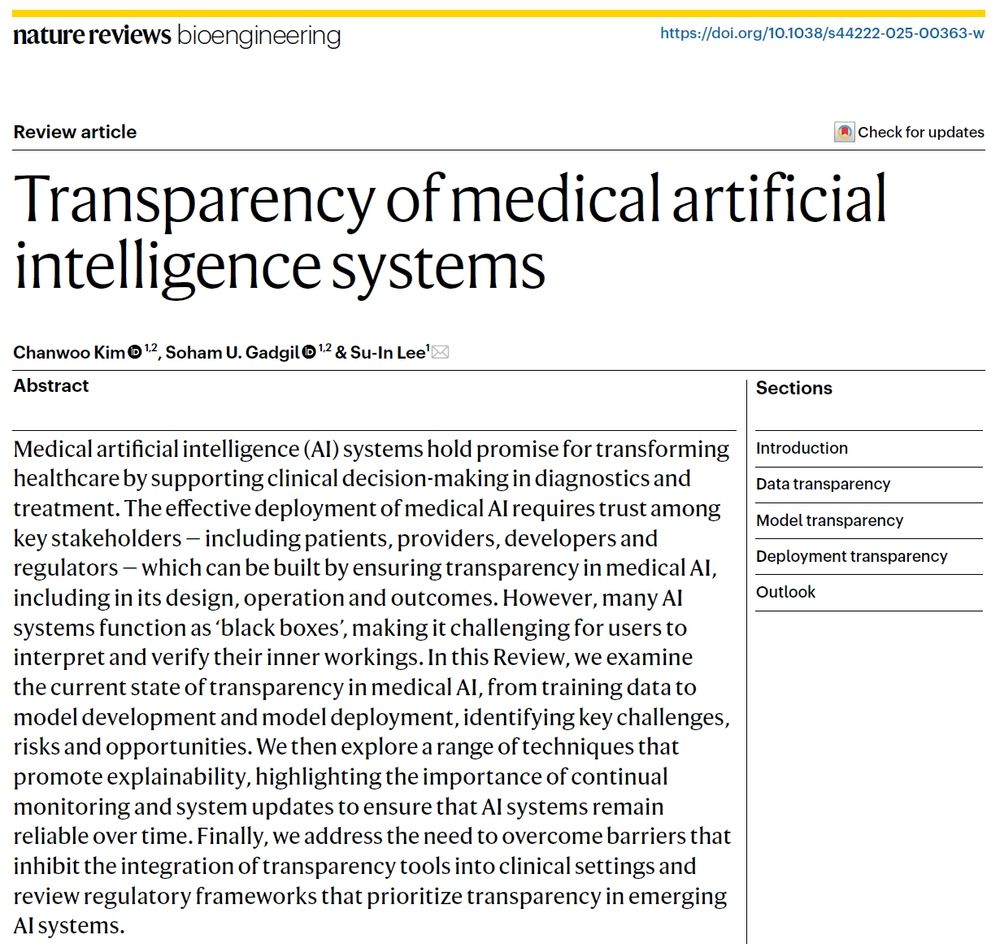
rdcu.be/eFnIc
Learn more: drive.google.com/file/d/15mMR...
#postdocjobs #AI #BiomedicalScience


Learn more: drive.google.com/file/d/15mMR...
#postdocjobs #AI #BiomedicalScience
The world lost a giant.
A big bear of a man.
With a huge smile.
With love for everyone.
With energy that could power a room.
I loved everything about Atul.
I loved how he was always happy.
I loved how excited he was about science and helping people.

The world lost a giant.
A big bear of a man.
With a huge smile.
With love for everyone.
With energy that could power a room.
I loved everything about Atul.
I loved how he was always happy.
I loved how excited he was about science and helping people.
@suinlee.bsky.social
and Scott Lundberg came out in 2017 and has 34k citations! Congratulations
@suinlee.bsky.social
and your team, what an achievement :-). arxiv.org/abs/1705.07874

@suinlee.bsky.social
and Scott Lundberg came out in 2017 and has 34k citations! Congratulations
@suinlee.bsky.social
and your team, what an achievement :-). arxiv.org/abs/1705.07874
More info here:
blog.iclr.cc/2025/04/23/i...
You can find all the sessions on the ICLR.cc schedule!

More info here:
blog.iclr.cc/2025/04/23/i...
You can find all the sessions on the ICLR.cc schedule!



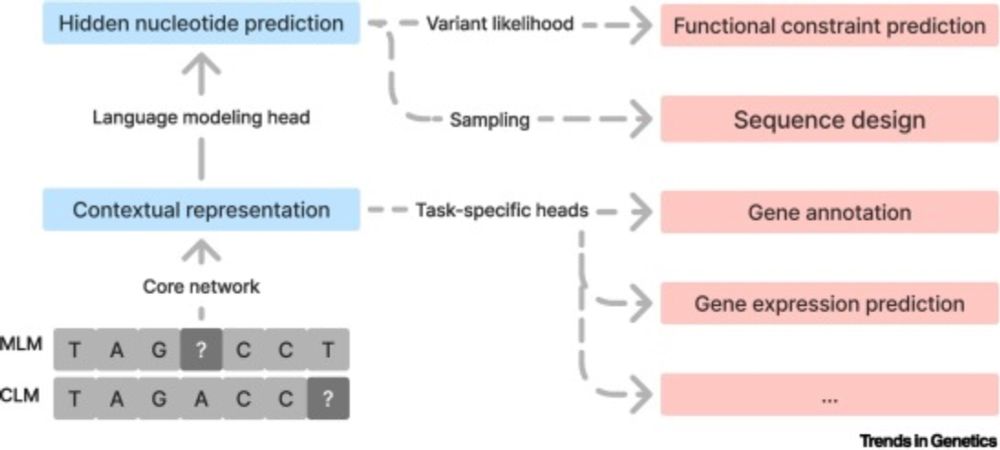

“Su-In’s body of work has completely transformed how people analyze and interpret AI/ML models. Her SHAP approach is used by essentially the entire field, not only in computational biology but all of computer science...”
news.cs.washington.edu/2025/04/02/s...

“Su-In’s body of work has completely transformed how people analyze and interpret AI/ML models. Her SHAP approach is used by essentially the entire field, not only in computational biology but all of computer science...”
news.cs.washington.edu/2025/04/02/s...


www.iscb.org/iscb-news-it...
www.iscb.org/iscb-news-it...

Check out our paper here: www.biorxiv.org/content/10.1...
Check out our paper here: www.biorxiv.org/content/10.1...
Check out our paper here: www.biorxiv.org/content/10.1...
Congrats: Anna, @xinmingtu.bsky.social , @lxsasse.bsky.social

Congrats: Anna, @xinmingtu.bsky.social , @lxsasse.bsky.social
Please join us in congratulating each of our 2025 awardees!


💡Theis is honored for his pioneering research in #ComputationalBiology.
👉More in our news:
t1p.de/u8olc
@fabiantheis.bsky.social @iscb.bsky.social
#AI #MachineLearning #ISCBInnovatorAward #CellResearch

💡Theis is honored for his pioneering research in #ComputationalBiology.
👉More in our news:
t1p.de/u8olc
@fabiantheis.bsky.social @iscb.bsky.social
#AI #MachineLearning #ISCBInnovatorAward #CellResearch



