


Held right at @embl.org Heidelberg, where I work!
4 days full of great science and great people. Fantastic meeting! 🔬
Curious to see where the single molecule field is going next! 👀
#EMBLSingleMolecule @events.embl.org

Held right at @embl.org Heidelberg, where I work!
4 days full of great science and great people. Fantastic meeting! 🔬
Curious to see where the single molecule field is going next! 👀
#EMBLSingleMolecule @events.embl.org
Thank you to all who visited and engaged in exciting discussions.
Grateful to the organizers for an excellent event! #EMBLSingleMolecule


Thank you to all who visited and engaged in exciting discussions.
Grateful to the organizers for an excellent event! #EMBLSingleMolecule


Panel 1️⃣: Single molecule genomics data analysis
Chair: @stirlingchurchman.bsky.social
Panel 2️⃣: Single molecule microscopy data analysis
Chairs: @laghalab.bsky.social, Dan Larson
@arnaudkr.bsky.social




Panel 1️⃣: Single molecule genomics data analysis
Chair: @stirlingchurchman.bsky.social
Panel 2️⃣: Single molecule microscopy data analysis
Chairs: @laghalab.bsky.social, Dan Larson
@arnaudkr.bsky.social
@arnaudkr.bsky.social @embl.org




From single-molecule insights to lively hallway conversations, it's clear: breakthroughs happen both in the lab and over lattes. ☕🧬
#EMBLSingleMolecule




From single-molecule insights to lively hallway conversations, it's clear: breakthroughs happen both in the lab and over lattes. ☕🧬
#EMBLSingleMolecule
www.nature.com/articles/d41...
www.nejm.org/doi/full/10....
www.nejm.org/doi/full/10....
www.nejm.org/doi/full/10....
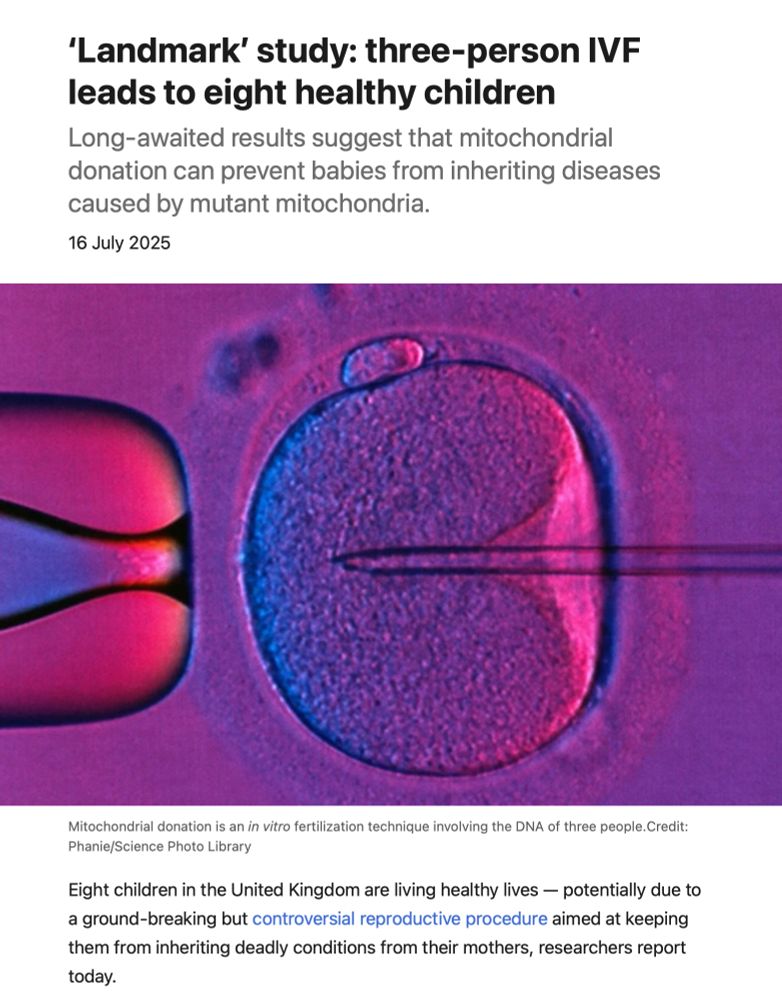
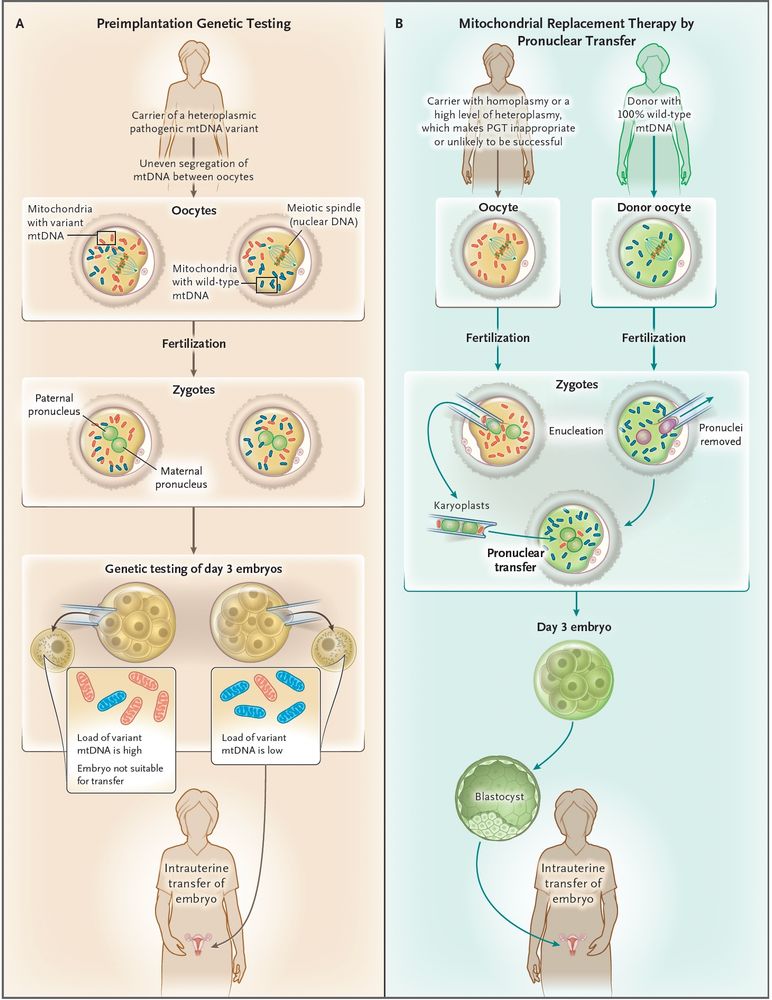
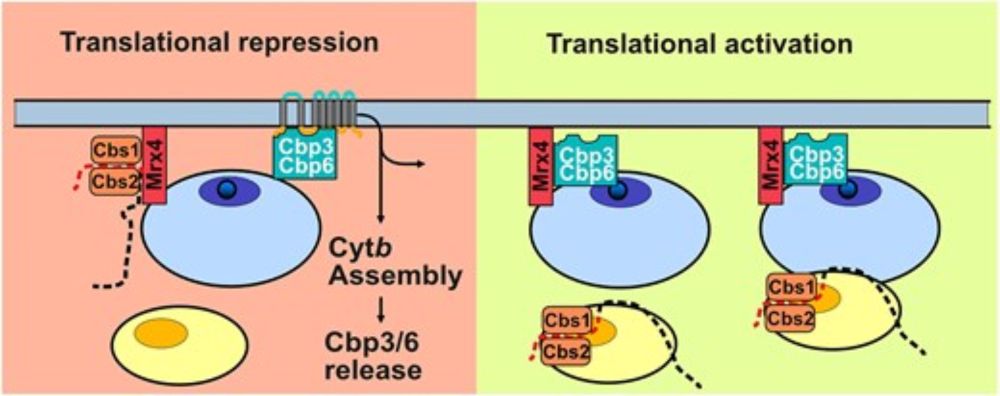
www.nature.com/articles/d41...
www.nejm.org/doi/full/10....
www.nejm.org/doi/full/10....
www.nejm.org/doi/full/10....
Most of the participants are already on site – now time for some science!
Our Scientific Organisers:
🔹 @stirlingchurchman.bsky.social
🔹@arnaudkr.bsky.social
🔹 @laghalab.bsky.social
🔹 Daniel Larson




Most of the participants are already on site – now time for some science!
Our Scientific Organisers:
🔹 @stirlingchurchman.bsky.social
🔹@arnaudkr.bsky.social
🔹 @laghalab.bsky.social
🔹 Daniel Larson
Fiber-seq is an LRS assay that maps chromatin accessibility, DNAme, and DNA sequence on single molecules.
If your jam is functional genomics/epigenetics, check this out.
We’re inviting a limited group of early users to try Fiber-seq at no cost.
Interested? Learn more 👉 explore.epicypher.com/fiber-seq-su...

Fiber-seq is an LRS assay that maps chromatin accessibility, DNAme, and DNA sequence on single molecules.
If your jam is functional genomics/epigenetics, check this out.

