Marcus Stoiber
@stoibs11.bsky.social
160 followers
160 following
11 posts
Associate director of modified base machine learning @nanopore | dad of 3 | amateur farmer | Nationals fan | views are my own
Posts
Media
Videos
Starter Packs
Reposted by Marcus Stoiber
Reposted by Marcus Stoiber
Marcus Stoiber
@stoibs11.bsky.social
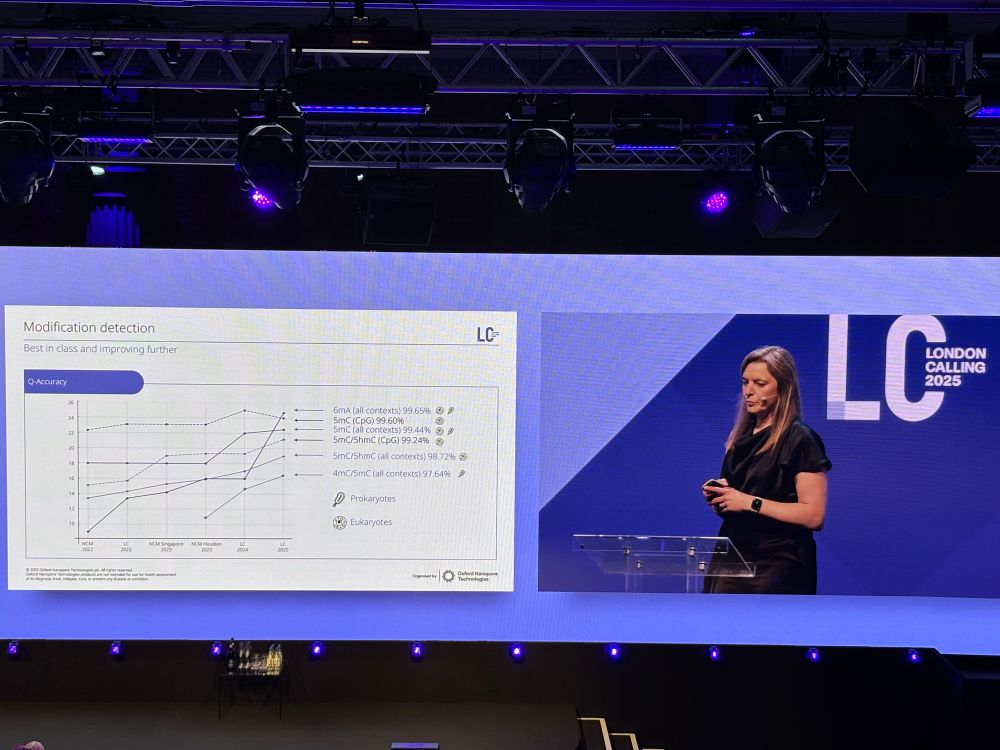
· May 21
Marcus Stoiber
@stoibs11.bsky.social
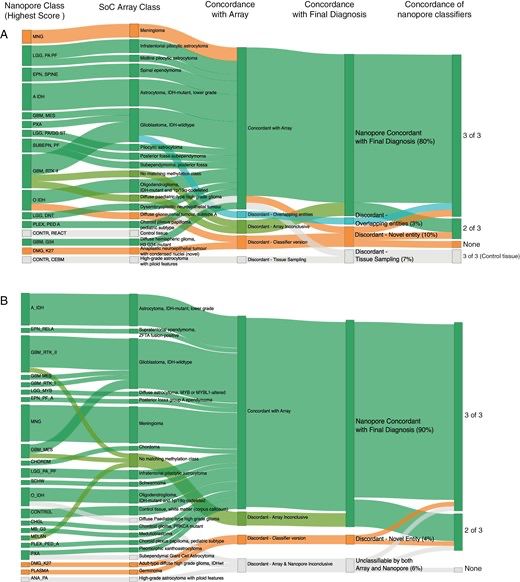
· Apr 2
Reposted by Marcus Stoiber
Reposted by Marcus Stoiber
Reposted by Marcus Stoiber
Reposted by Marcus Stoiber
Reposted by Marcus Stoiber