at Broad Institute | Former PhD student at Sanger Institute | Lineage tracing, cancer genomics, human development
Find out how Tim’s group is exploring using large-scale single-cell and spatial data to trace cell lineages, understand cancer origins, and uncover how mutations drive disease.
www.ebi.ac.uk/about/news/p...
🖥️🧬

Find out how Tim’s group is exploring using large-scale single-cell and spatial data to trace cell lineages, understand cancer origins, and uncover how mutations drive disease.
www.ebi.ac.uk/about/news/p...
🖥️🧬
@emblebi.bsky.social!
Reach out if interested or you have any questions, and apply here: embl.wd103.myworkdayjobs.com/en-US/EMBL/j...

@emblebi.bsky.social!
Reach out if interested or you have any questions, and apply here: embl.wd103.myworkdayjobs.com/en-US/EMBL/j...

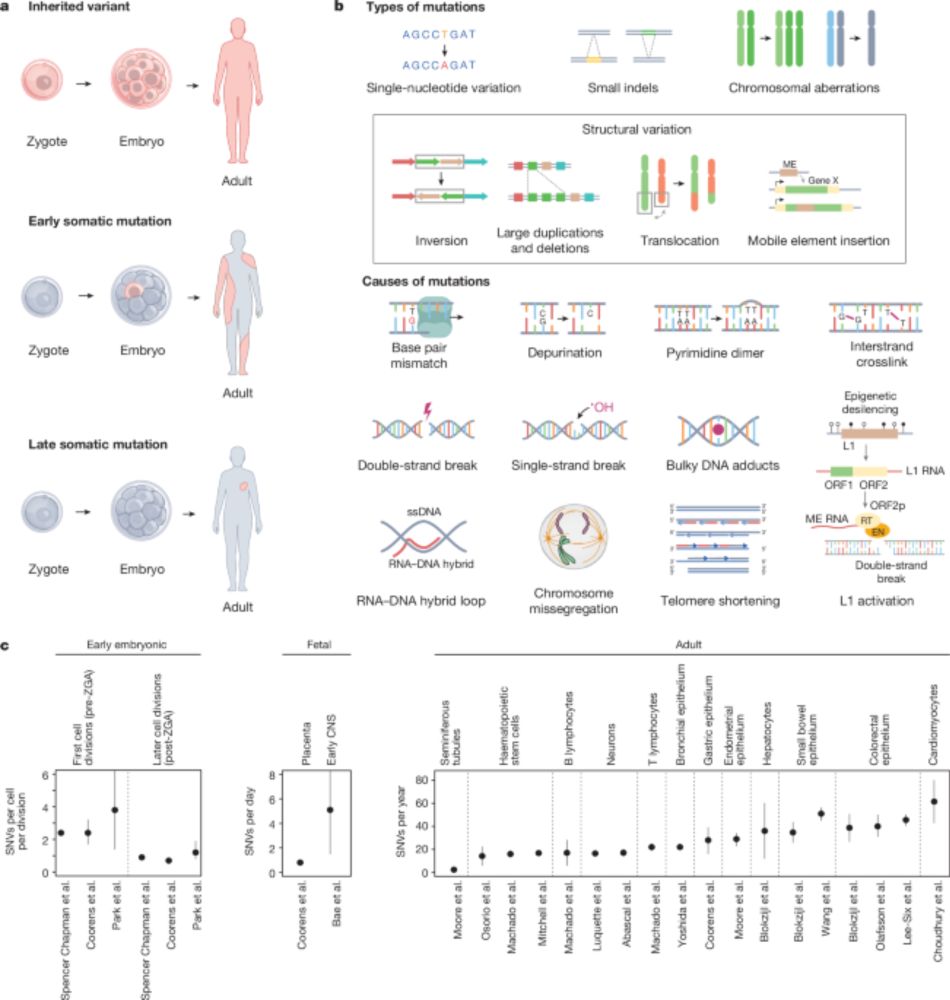
🗣 Dr. @timcoorens.bsky.social from @ebi.embl.org :
🧪 "Somatic mutations across normal tissues: past, present, future"
📅 June 30, 1 PM
📍 Theatre room, CiMUS
#RedeCiGUS #FondosEuropeos
@mobilegenomes.bsky.social @usc.gal

🗣 Dr. @timcoorens.bsky.social from @ebi.embl.org :
🧪 "Somatic mutations across normal tissues: past, present, future"
📅 June 30, 1 PM
📍 Theatre room, CiMUS
#RedeCiGUS #FondosEuropeos
@mobilegenomes.bsky.social @usc.gal
Check out our approach to modernise chemotherapy treatment published today in @natgenet.nature.com. From @cniostopcancer.bsky.social #TailorBio @cruk-ci.bsky.social www.nature.com/articles/s41... More details 👇
Check out our approach to modernise chemotherapy treatment published today in @natgenet.nature.com. From @cniostopcancer.bsky.social #TailorBio @cruk-ci.bsky.social www.nature.com/articles/s41... More details 👇
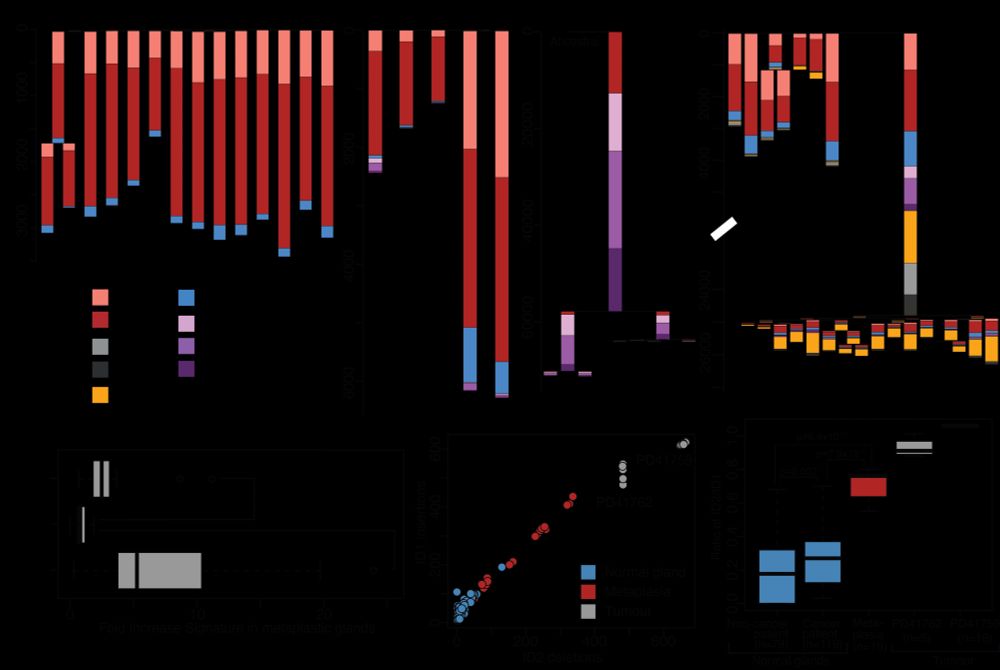
Please check out my new perspective on metastasis evolution in @natrevcancer.nature.com if you need a reprieve from the news.
I tried my best to develop some new conceptual thoughts here, you be the judge whether I succeeded.
Naxerova explains how genetic analysis of primary tumors & metastases can distinguish competing metastasis models, arguing that quantifying whether metastasis occurs through nonrandom lineage selection could offer insights into metastasis biology.
📖 👇
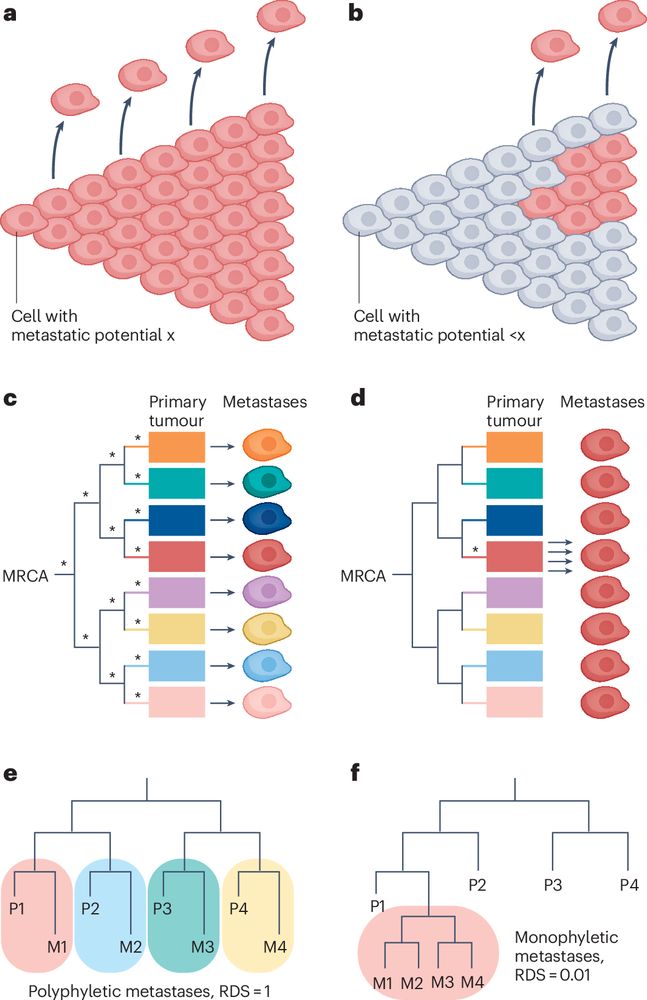
Please check out my new perspective on metastasis evolution in @natrevcancer.nature.com if you need a reprieve from the news.
I tried my best to develop some new conceptual thoughts here, you be the judge whether I succeeded.

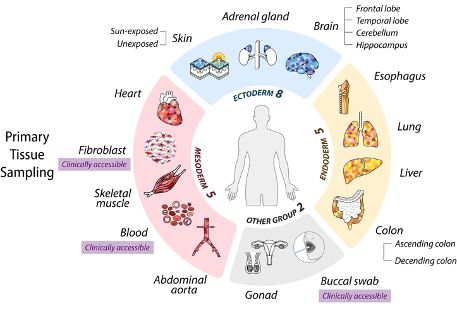
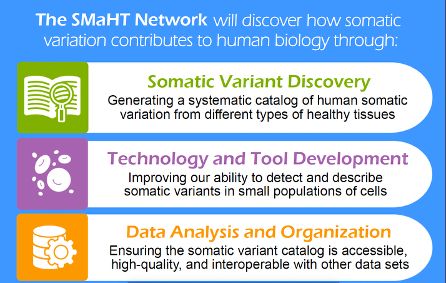
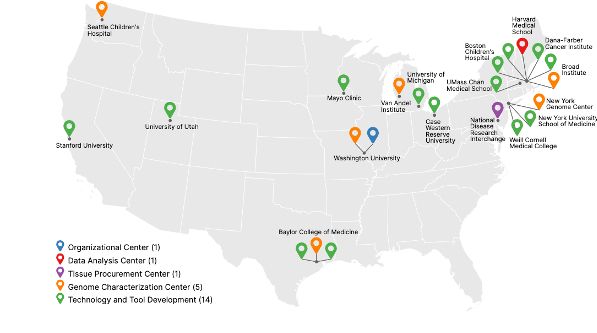
Our Linktree includes all SMaHT websites and social media accounts for easy and consistent access.
Find all our social media accounts in one place:
linktr.ee/smahtnetwork
Our Linktree includes all SMaHT websites and social media accounts for easy and consistent access.
Find all our social media accounts in one place:
linktr.ee/smahtnetwork
#Genomics #SomaticEvolution
#Genomics #SomaticEvolution
www.nature.com/articles/s41...
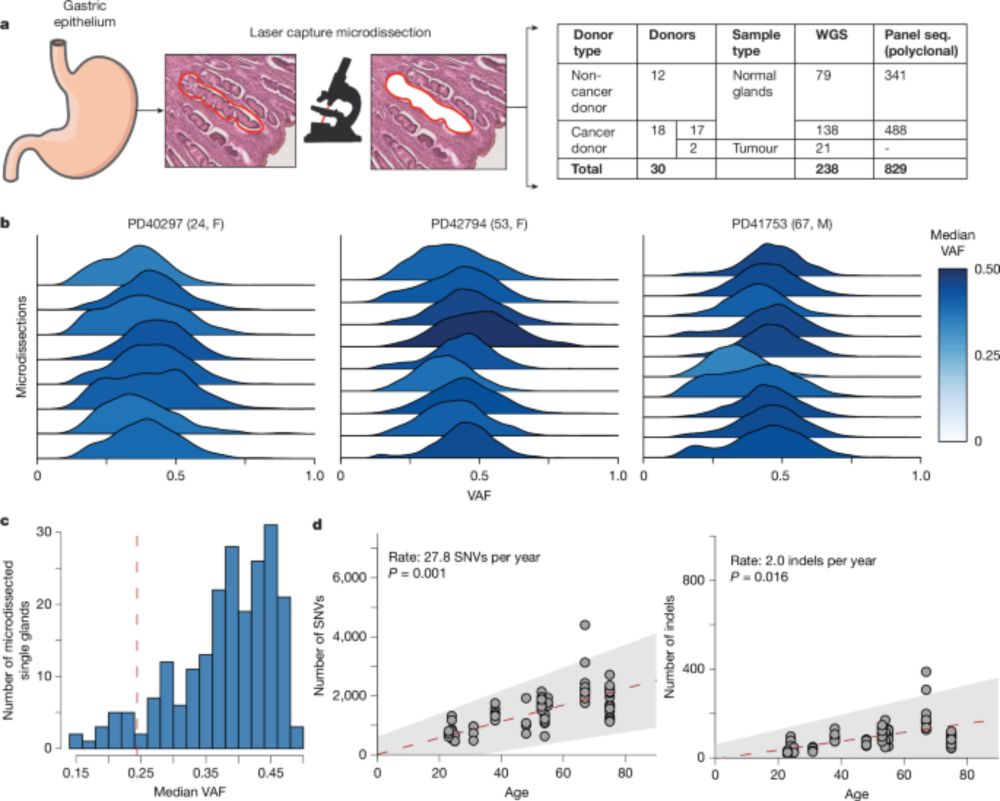
www.nature.com/articles/s41...