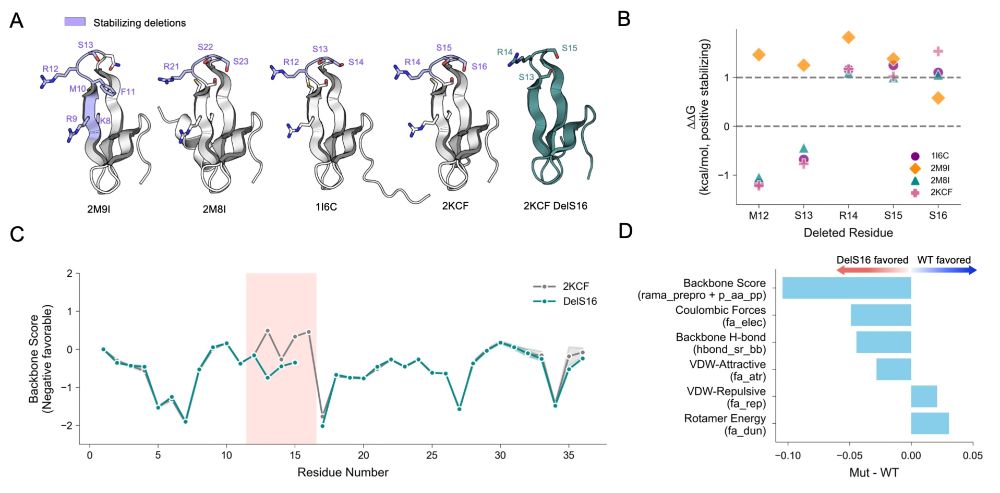
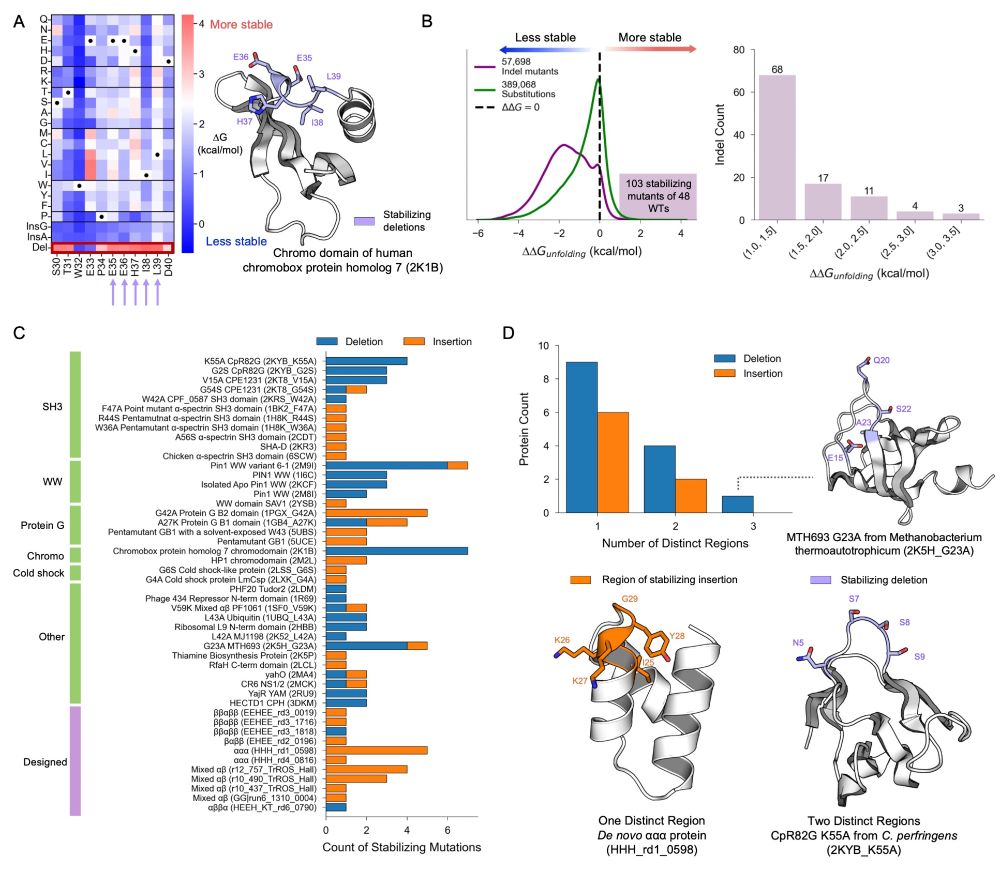
Now more than ever before, it is critical that we bridge the gap btw scientists & the community to rebuild trust & address misinformation. Join the national #McClintockLetters movement to #DefendResearch, improve #SciComm, & share your scientific story! ⤵️

Now more than ever before, it is critical that we bridge the gap btw scientists & the community to rebuild trust & address misinformation. Join the national #McClintockLetters movement to #DefendResearch, improve #SciComm, & share your scientific story! ⤵️
We know proteins fluctuate between different conformations- but by how much? How does it vary from protein to protein? Can highly stable domains have low stability segments? @ajrferrari.bsky.social experimentally tested >5,000 domains to find out!
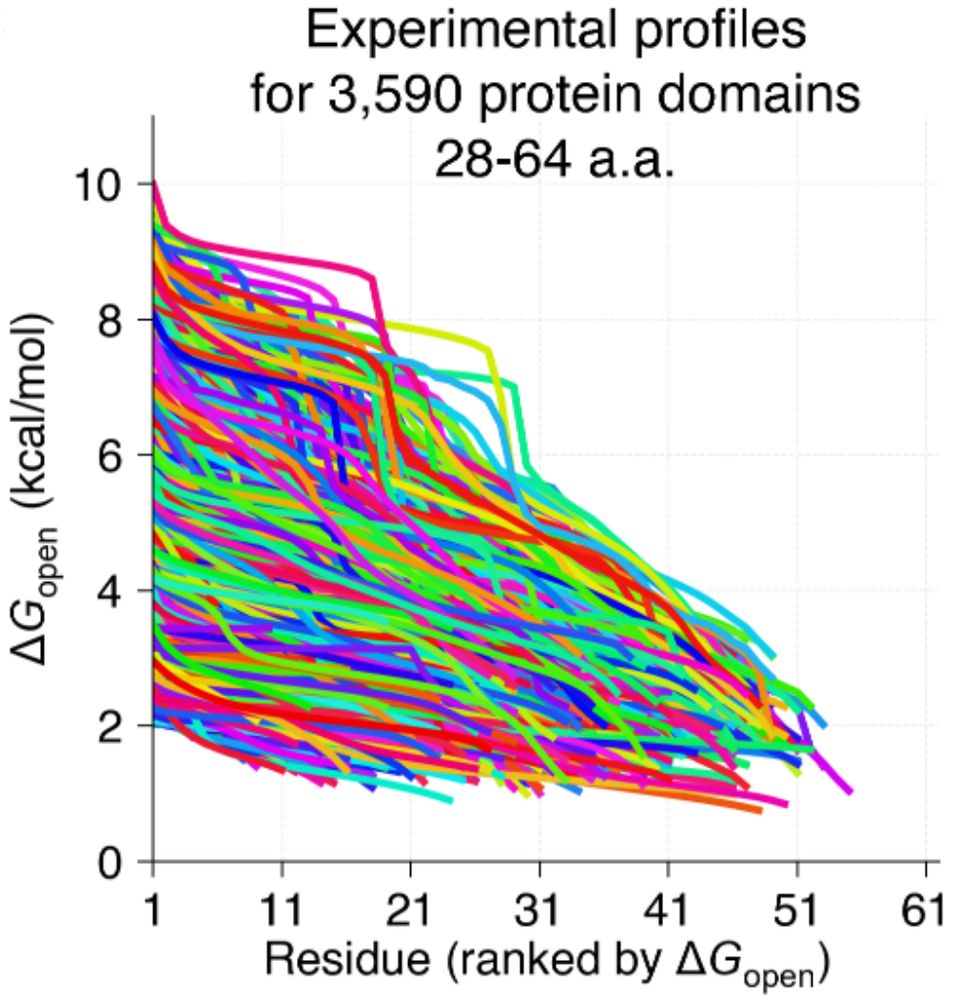
We know proteins fluctuate between different conformations- but by how much? How does it vary from protein to protein? Can highly stable domains have low stability segments? @ajrferrari.bsky.social experimentally tested >5,000 domains to find out!