Alexander Belyy
@alexanderbelyy.bsky.social
210 followers
310 following
20 posts
Assistant professor, University of Groningen, NL.
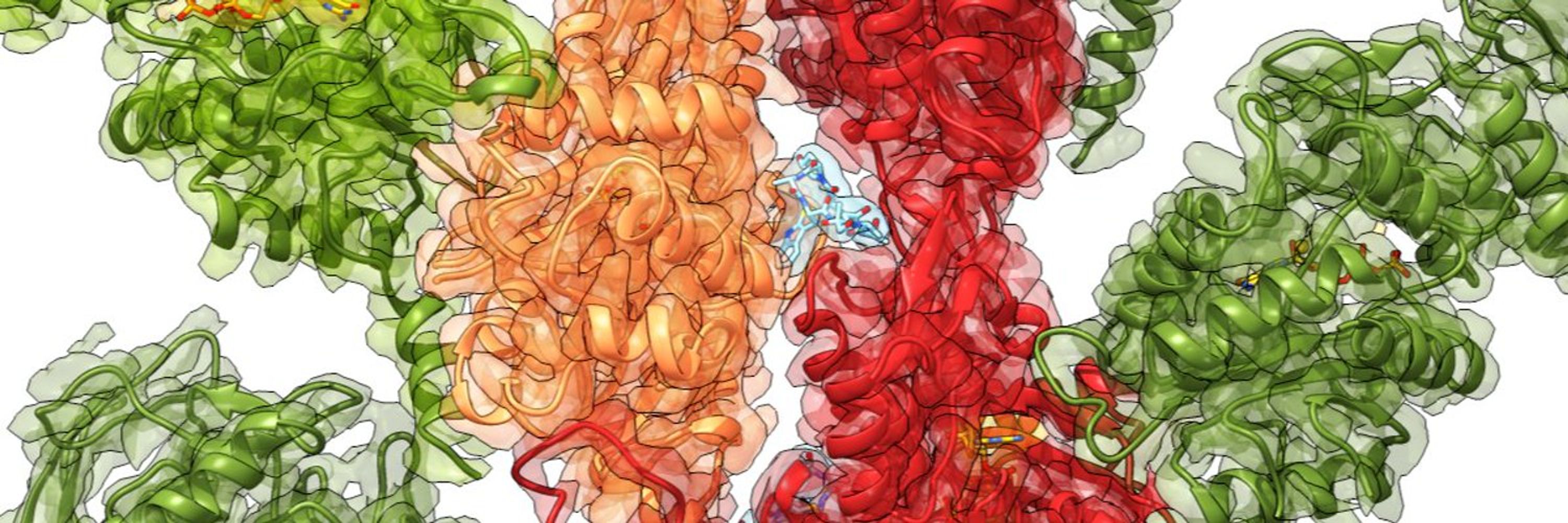
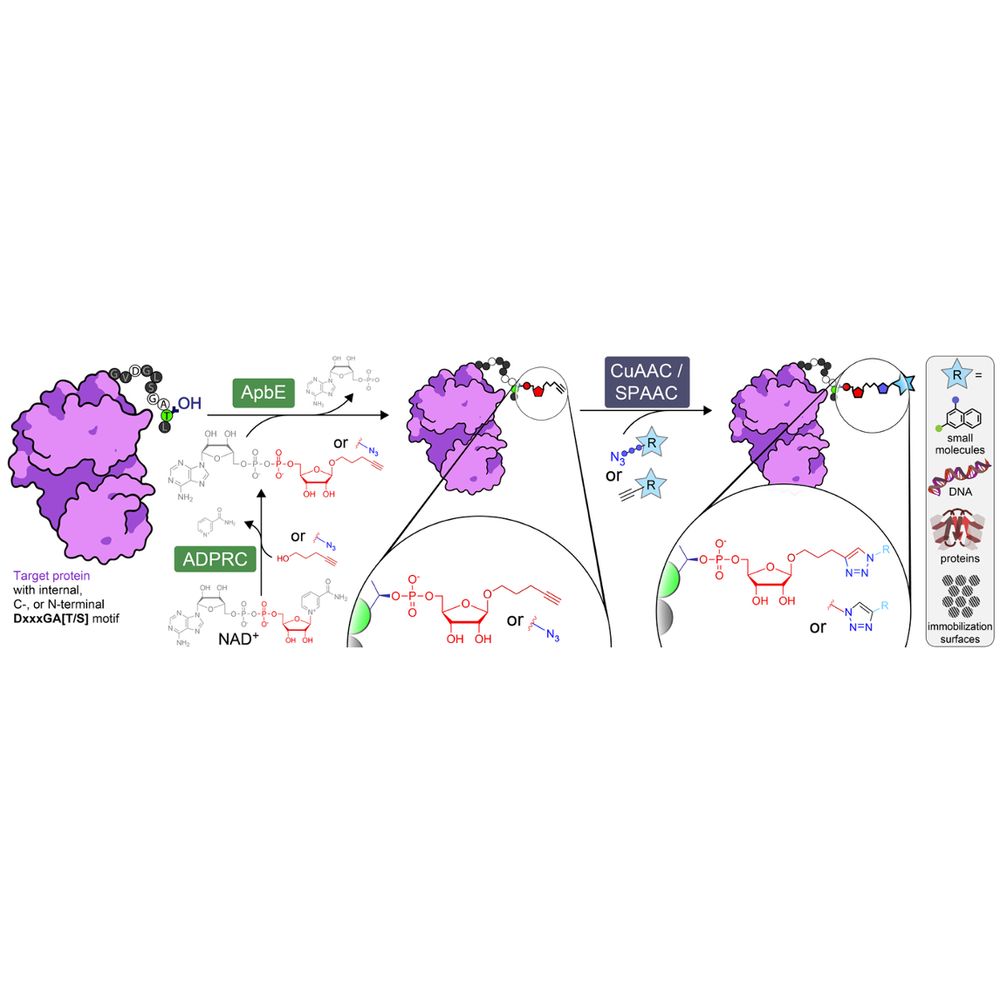
Studying bacterial toxins and actin cytoskeleton using cryo-EM.
Posts
Media
Videos
Starter Packs
Reposted by Alexander Belyy
Reposted by Alexander Belyy
Reposted by Alexander Belyy
Reposted by Alexander Belyy
Kasia Tych
@biophysk.bsky.social
· Jan 30
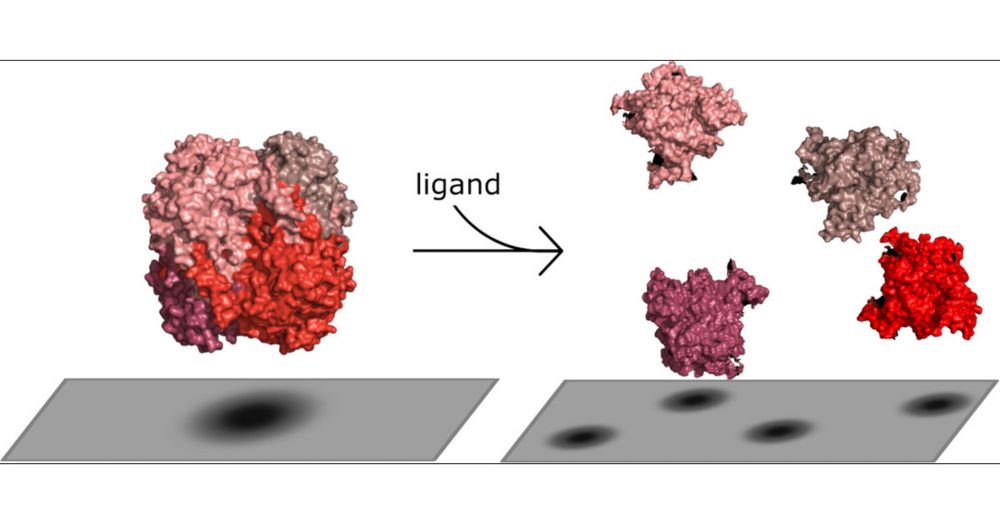
Impact of Ligand-Induced Oligomer Dissociation on Enzyme Diffusion, Directly Observed at the Single-Molecule Level
The existence of the phenomenon of enhanced enzyme diffusion (EED) has been a topic of debate in recent literature. One proposed mechanism to explain the origin of EED is oligomeric enzyme dissociation. We used mass photometry (MP), a label-free single-molecule technique, to investigate the dependence of the oligomeric states of several enzymes on their ligands. The studied enzymes of interest are catalase, aldolase, alkaline phosphatase, and vanillyl-alcohol oxidase (VAO). We compared the ratios of oligomeric states in the presence and absence of the substrate as well as different substrate and inhibitor concentrations. Catalase and aldolase were found to dissociate into smaller oligomers in the presence of their substrates, independently of inhibition, while for alkaline phosphatase and VAO, different behaviors were observed. Thus, we have identified a possible mechanism which explains the previously observed diffusion enhancement in vitro. This enhancement may occur due to the dissociation of oligomers through ligand binding.
pubs.acs.org