We fused TIRAP’s phosphoinositide-binding domain to caspase-11’s CARD. The aligned TIRAP-CARD bound LPS and aggregated. This supports electrostatic interactions as key for CARD-lipid binding. #Caspase11

We fused TIRAP’s phosphoinositide-binding domain to caspase-11’s CARD. The aligned TIRAP-CARD bound LPS and aggregated. This supports electrostatic interactions as key for CARD-lipid binding. #Caspase11
Caspase-11’s N-terminal positive charges suggest aggregation depends on negatively charged headgroups. PIP-strip assays confirmed binding to phosphorylated lipids, including PI(3,4,5)P3 expanding its ligand repertoire. #Caspase11 #Lipid

Caspase-11’s N-terminal positive charges suggest aggregation depends on negatively charged headgroups. PIP-strip assays confirmed binding to phosphorylated lipids, including PI(3,4,5)P3 expanding its ligand repertoire. #Caspase11 #Lipid
Introducing D20K/E29K into AcCARD converted it from a PGPC receptor to a dual PGPC/LPS receptor, mimicking caspase-11. These mutations enabled LPS-induced aggregation and trypsin
resistance #Caspase11 #ProteinEngineering
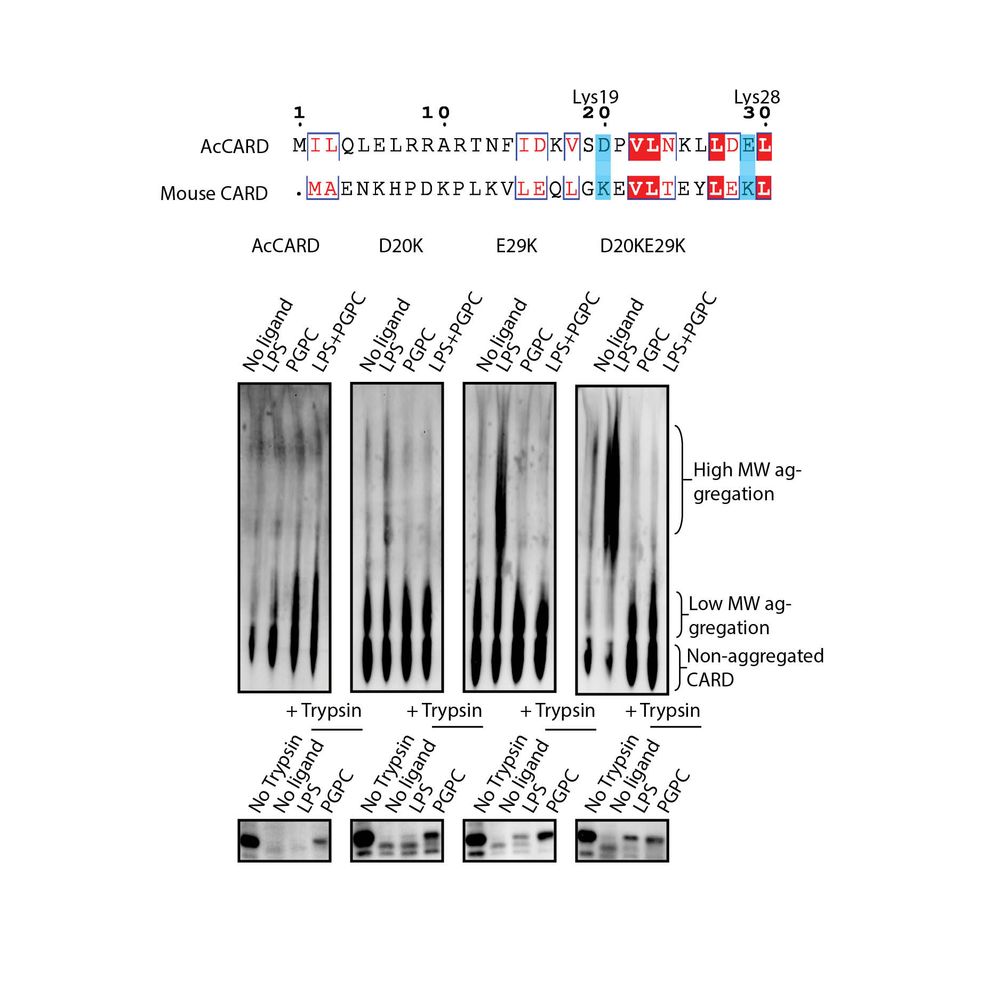
Introducing D20K/E29K into AcCARD converted it from a PGPC receptor to a dual PGPC/LPS receptor, mimicking caspase-11. These mutations enabled LPS-induced aggregation and trypsin
resistance #Caspase11 #ProteinEngineering
By fusing caspase-11’s N-terminal region to AcCARD’s PGPC-binding domain, we created Casp11-AcCARD, which gained LPS-induced aggregation and trypsin resistance. #Immunology #Caspase11 #ProteinEngineering

By fusing caspase-11’s N-terminal region to AcCARD’s PGPC-binding domain, we created Casp11-AcCARD, which gained LPS-induced aggregation and trypsin resistance. #Immunology #Caspase11 #ProteinEngineering
We identified AcCARD, a CARD-containing protein from Amphilophus citrinellus, which binds PGPC but not LPS. This supports the idea that CARD functions can be selectively engineered to recognize different ligands. #Caspase11
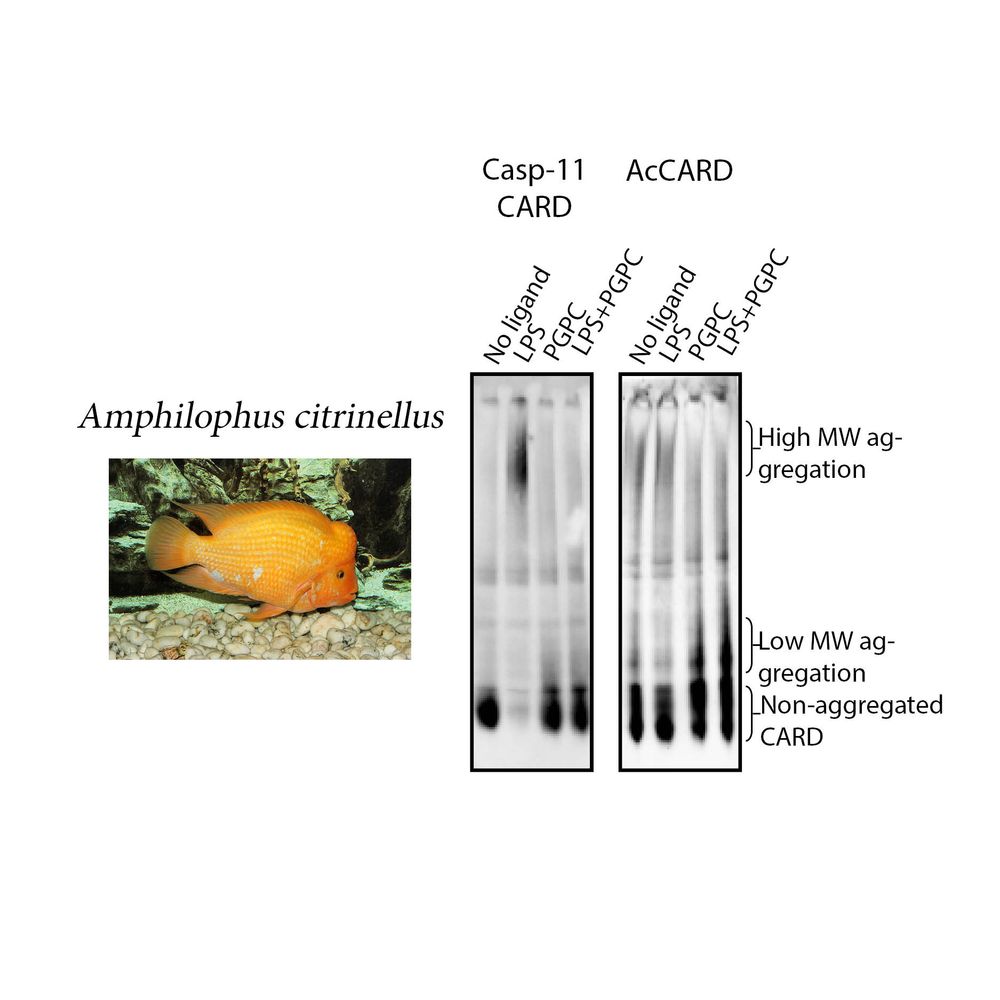
We identified AcCARD, a CARD-containing protein from Amphilophus citrinellus, which binds PGPC but not LPS. This supports the idea that CARD functions can be selectively engineered to recognize different ligands. #Caspase11
Caspase-11’s CARD uses distinct regions for lipid binding and oligomerization. Phe76 is crucial for binding both LPS and PGPC, but only LPS triggers aggregation. Deletions in residues 10–25 reveal key differences in how these lipids interact. #Caspase11

Caspase-11’s CARD uses distinct regions for lipid binding and oligomerization. Phe76 is crucial for binding both LPS and PGPC, but only LPS triggers aggregation. Deletions in residues 10–25 reveal key differences in how these lipids interact. #Caspase11
We reconstituted caspase-11-/- iBMDMs with WT, C254A, and F76E variants. While WT triggered IL-1β and LDH release, F76E showed impaired responses at low LPS doses, confirming Phe76’s role in LPS binding and signaling. #Caspase11

We reconstituted caspase-11-/- iBMDMs with WT, C254A, and F76E variants. While WT triggered IL-1β and LDH release, F76E showed impaired responses at low LPS doses, confirming Phe76’s role in LPS binding and signaling. #Caspase11
PGPC protected 16 of 22 caspase-4 CARDs. Phe45 and Phe76 were enriched in PGPC-protected CARDs, but some protected variants lacked both, suggesting additional binding determinants. #Immunology #Caspase4 #LipidBinding

PGPC protected 16 of 22 caspase-4 CARDs. Phe45 and Phe76 were enriched in PGPC-protected CARDs, but some protected variants lacked both, suggesting additional binding determinants. #Immunology #Caspase4 #LipidBinding
To examine the role of acyl chain length, we tested LysoPCs (6–18 carbons). Only LysoPCs with ≥14 carbons protected caspase-11. This protective effect also occurred with the CARD, similar to LPS and PGPC interactions. #Immunology #Caspase11 #LipidBinding

To examine the role of acyl chain length, we tested LysoPCs (6–18 carbons). Only LysoPCs with ≥14 carbons protected caspase-11. This protective effect also occurred with the CARD, similar to LPS and PGPC interactions. #Immunology #Caspase11 #LipidBinding
We tested 16-carbon monoacylated lipids with various negatively charged headgroups. Most protected caspase-11 from trypsin digestion, suggesting caspase-11 interacts broadly with monoacylated lipids.

We tested 16-carbon monoacylated lipids with various negatively charged headgroups. Most protected caspase-11 from trypsin digestion, suggesting caspase-11 interacts broadly with monoacylated lipids.
Mass spectrometry of caspase-11 after trypsin digestion showed that the 20 kDa fragment lacked peptides before K62, while the 35 kDa band retained the full N-terminal sequence. K62, located in the CARD, is protected upon lipid binding. #Caspase11

Mass spectrometry of caspase-11 after trypsin digestion showed that the 20 kDa fragment lacked peptides before K62, while the 35 kDa band retained the full N-terminal sequence. K62, located in the CARD, is protected upon lipid binding. #Caspase11
Unbound caspase-11 degrades to a 20 kDa fragment within 15 min. However, LPS-EB, LPS-RS, and PGPC induce a trypsin-resistant state, yielding a 35 kDa band, suggesting ligand binding alters caspase-11 conformation. #ProteinStructure #LipidBinding

Unbound caspase-11 degrades to a 20 kDa fragment within 15 min. However, LPS-EB, LPS-RS, and PGPC induce a trypsin-resistant state, yielding a 35 kDa band, suggesting ligand binding alters caspase-11 conformation. #ProteinStructure #LipidBinding
CD analysis suggested caspase-11-lipid interactions alter protein conformation, influencing protease accessibility. Limited trypsin digestion revealed ligand-dependent cleavage patterns, detected via immunoblotting. #Immunology #Caspase11

CD analysis suggested caspase-11-lipid interactions alter protein conformation, influencing protease accessibility. Limited trypsin digestion revealed ligand-dependent cleavage patterns, detected via immunoblotting. #Immunology #Caspase11
CD analysis revealed a shift toward a more α-helical conformation upon binding non-aggregating ligands LPS-RS and PGPC. This suggests distinct ligand-induced structural changes. #Immunology #Caspase11 #ProteinStructure

CD analysis revealed a shift toward a more α-helical conformation upon binding non-aggregating ligands LPS-RS and PGPC. This suggests distinct ligand-induced structural changes. #Immunology #Caspase11 #ProteinStructure
Lipid size and properties affect caspase-11 aggregation. PGPC, a small monoacylated lipid from oxPAPC, doesn’t aggregate caspase-11 but blocks LPS-induced aggregation, suggesting direct interaction. #Immunology #LPS

Lipid size and properties affect caspase-11 aggregation. PGPC, a small monoacylated lipid from oxPAPC, doesn’t aggregate caspase-11 but blocks LPS-induced aggregation, suggesting direct interaction. #Immunology #LPS
Caspase-11 aggregation is often used as a proxy for LPS binding, but not all LPS that bind caspase-11 induce aggregation. For instance, LPS from Rhodobacter sphaeroides doesn’t trigger aggregation but inhibits E. coli LPS-induced aggregation.

Caspase-11 aggregation is often used as a proxy for LPS binding, but not all LPS that bind caspase-11 induce aggregation. For instance, LPS from Rhodobacter sphaeroides doesn’t trigger aggregation but inhibits E. coli LPS-induced aggregation.

