


@roman-bushuiev.bsky.social for receiving the Google PhD Fellowship 2025 in Health Research! 🎉💰 goo.gle/43wJWw8
@roman-bushuiev.bsky.social for receiving the Google PhD Fellowship 2025 in Health Research! 🎉💰 goo.gle/43wJWw8

www.nature.com/articles/s41...
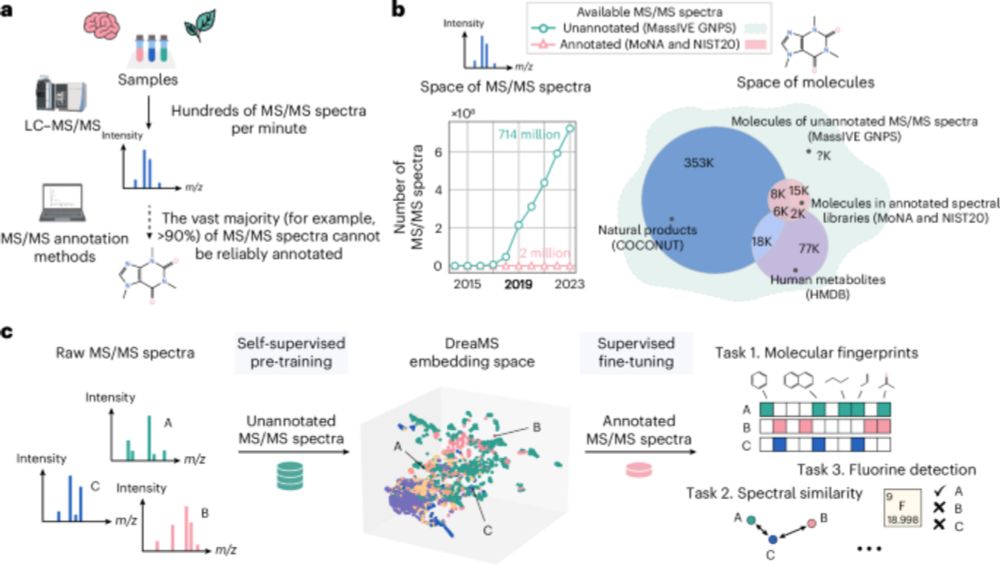
www.nature.com/articles/s41...



Two new machine learning models achieve up to a 300% improvement in de novo molecular generation given mass spectra and corresponding chemical formulae. 🔥 1/n

Two new machine learning models achieve up to a 300% improvement in de novo molecular generation given mass spectra and corresponding chemical formulae. 🔥 1/n
@roman-bushuiev.bsky.social
@anton-bushuiev.bsky.social
@josef-sivic.bsky.social
@pluskal-lab.org
NeurIPS 2024 paper: arxiv.org/abs/2410.23326
#ChemSky #MassSpec #AI4Science

@roman-bushuiev.bsky.social
@anton-bushuiev.bsky.social
@josef-sivic.bsky.social
@pluskal-lab.org
NeurIPS 2024 paper: arxiv.org/abs/2410.23326
#ChemSky #MassSpec #AI4Science

🛡️The dataset is now certified on Polaris! polarishub.io/datasets/rom...
youtu.be/G8ZnVRm0ogc

🛡️The dataset is now certified on Polaris! polarishub.io/datasets/rom...
youtu.be/G8ZnVRm0ogc
Adamo Young,
@roman-bushuiev.bsky.social,
@anton-bushuiev.bsky.social,
Raman Samusevich

Adamo Young,
@roman-bushuiev.bsky.social,
@anton-bushuiev.bsky.social,
Raman Samusevich


We’ll be at the conference doing short interviews with researchers and handing out some Polaris merch!
Here’s who we have on the shortlist. 🧵

We’ll be at the conference doing short interviews with researchers and handing out some Polaris merch!
Here’s who we have on the shortlist. 🧵
This may be the most carefully documented dataset on @polarishq.bsky.social. Great work by @roman-bushuiev.bsky.social!
Do I have any #MassSpec researchers in my 🦋 network yet? I would love to hear what you think! github.com/polaris-hub/...
#mass #spectrometry #dataset #benchmark

This may be the most carefully documented dataset on @polarishq.bsky.social. Great work by @roman-bushuiev.bsky.social!
Do I have any #MassSpec researchers in my 🦋 network yet? I would love to hear what you think! github.com/polaris-hub/...
#mass #spectrometry #dataset #benchmark

