Archaeological Proteomics Lab
@archaeoprotein.bsky.social
690 followers
150 following
8 posts
Lab at University of Reading using #archsci #Palaeoproteomics #ZooMS and #Zooarchaeology 🦌🦏🦴🐂 to study past human behaviour; home to project COEXIST. PI Karen Ruebens.
Posts
Media
Videos
Starter Packs
Reposted by Archaeological Proteomics Lab
Reposted by Archaeological Proteomics Lab
Reposted by Archaeological Proteomics Lab
Reposted by Archaeological Proteomics Lab
Reposted by Archaeological Proteomics Lab
Takumi Tsutaya
@tsutatsuta.bsky.social
· Apr 10
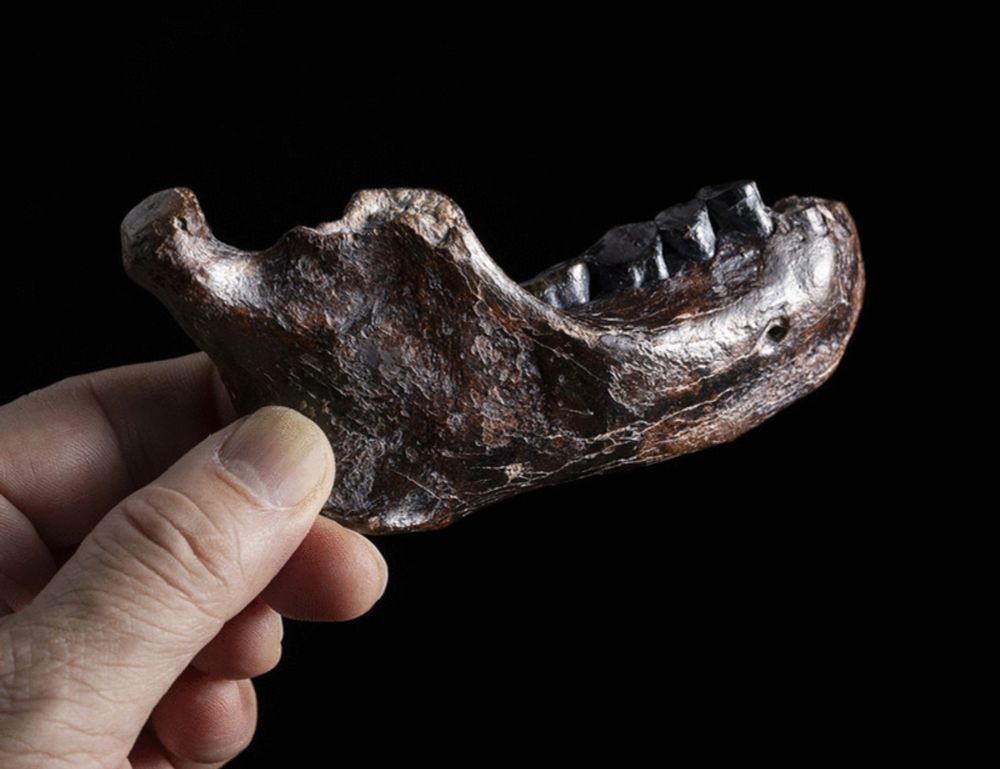
A male Denisovan mandible from Pleistocene Taiwan
Denisovans are an extinct hominin group defined by ancient genomes of Middle to Late Pleistocene fossils from southern Siberia. Although genomic evidence suggests their widespread distribution through...
doi.org
Reposted by Archaeological Proteomics Lab
Reposted by Archaeological Proteomics Lab
Reposted by Archaeological Proteomics Lab
Reposted by Archaeological Proteomics Lab
Reposted by Archaeological Proteomics Lab
Reposted by Archaeological Proteomics Lab
Reposted by Archaeological Proteomics Lab
Reposted by Archaeological Proteomics Lab