




🏆 Generalist Prize: Team @altos_labs ranked highest across 7 metrics. They showed the most reliable generalization—robust performance across diverse criteria vs. optimization for a single score.

🏆 Generalist Prize: Team @altos_labs ranked highest across 7 metrics. They showed the most reliable generalization—robust performance across diverse criteria vs. optimization for a single score.


Their key insight? Pure AI approaches didn't beat statistical baselines so they integrated both.

Their key insight? Pure AI approaches didn't beat statistical baselines so they integrated both.
Over 5,000 participants from 114 countries competed to build AI models that predict cellular responses to genetic perturbations. Today we're announcing the winners and reflecting on what we learned.

Over 5,000 participants from 114 countries competed to build AI models that predict cellular responses to genetic perturbations. Today we're announcing the winners and reflecting on what we learned.
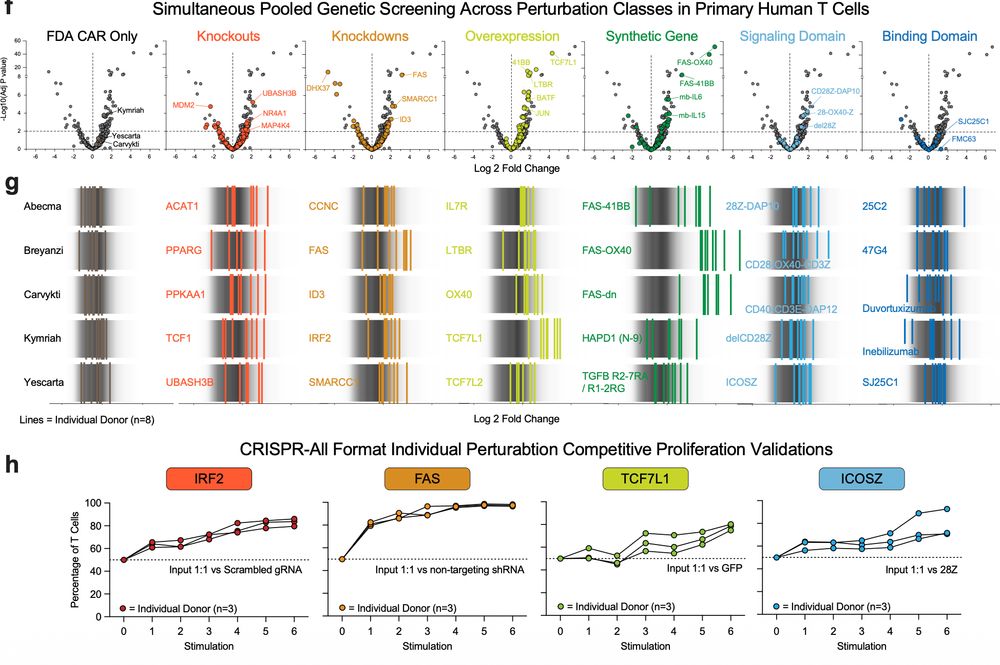
Evaluating them in matched conditions allowed the team to precisely identify which edits enhance function under chronic stimulation.

Evaluating them in matched conditions allowed the team to precisely identify which edits enhance function under chronic stimulation.



Evo was able to produce novel proteins that inhibited SpCas9 and improved phage survival, demonstrating its ability to propose functional products in systems where sequence-based approaches often fail.

Evo was able to produce novel proteins that inhibited SpCas9 and improved phage survival, demonstrating its ability to propose functional products in systems where sequence-based approaches often fail.
Prompted on the genomic contexts typical of these pairs, Evo successfully generated a range of diverse candidates, including several experimentally confirmed toxins and antitoxins.

Prompted on the genomic contexts typical of these pairs, Evo successfully generated a range of diverse candidates, including several experimentally confirmed toxins and antitoxins.
In sequence-recovery tests, Evo reliably reconstructed conserved genes from partial inputs, with accuracy improving across model versions.

In sequence-recovery tests, Evo reliably reconstructed conserved genes from partial inputs, with accuracy improving across model versions.

His work solves a decades-old mystery in cell biology, revealing the enzyme responsible for BMP synthesis: www.aaas.org/news/researc...

His work solves a decades-old mystery in cell biology, revealing the enzyme responsible for BMP synthesis: www.aaas.org/news/researc...
This means researchers can now choose variants optimized for maximum efficiency, specificity, or a balance of both.

This means researchers can now choose variants optimized for maximum efficiency, specificity, or a balance of both.


Existing recombinases, however, have limitations–managing only ~5% efficiency & often hitting hundreds of off-target sites.

Existing recombinases, however, have limitations–managing only ~5% efficiency & often hitting hundreds of off-target sites.


