Ariel Kaplan
@arielkaplan.bsky.social
990 followers
220 following
41 posts
Single molecule biophysics, Optical Tweezers, Chromatin, Transcription, Polymerases, Helicases
Associate Professor, Technion-Israel Institute of Technology
https://kaplan.net.technion.ac.il/
Posts
Media
Videos
Starter Packs
Pinned
Ariel Kaplan
@arielkaplan.bsky.social
· May 27

Intrinsically disordered regions facilitate Msn2 target search to drive promoter selectivity
Transcription factors (TFs) regulate gene expression by binding specific DNA motifs, yet only a fraction of putative sites is occupied in vivo. Intrinsically disordered regions (IDRs) have emerged as ...
www.biorxiv.org
Ariel Kaplan
@arielkaplan.bsky.social
· Jul 23
Reposted by Ariel Kaplan
Reposted by Ariel Kaplan
BejaLab
@bejalab.bsky.social
· May 31
BejaLab
@bejalab.bsky.social
· May 30
Marine Asgard Archaea's Light-Harvesting Rhodopsin Revealed
In a groundbreaking advancement that challenges existing paradigms of microbial photobiology, researchers have unveiled novel structural insights into how marine Asgard archaea harness light energy.
scienmag.com
Reposted by Ariel Kaplan
Reposted by Ariel Kaplan
Reposted by Ariel Kaplan
Alexis Verger 🧬🧫🧪
@alexis-verger.cpesr.fr
· May 29
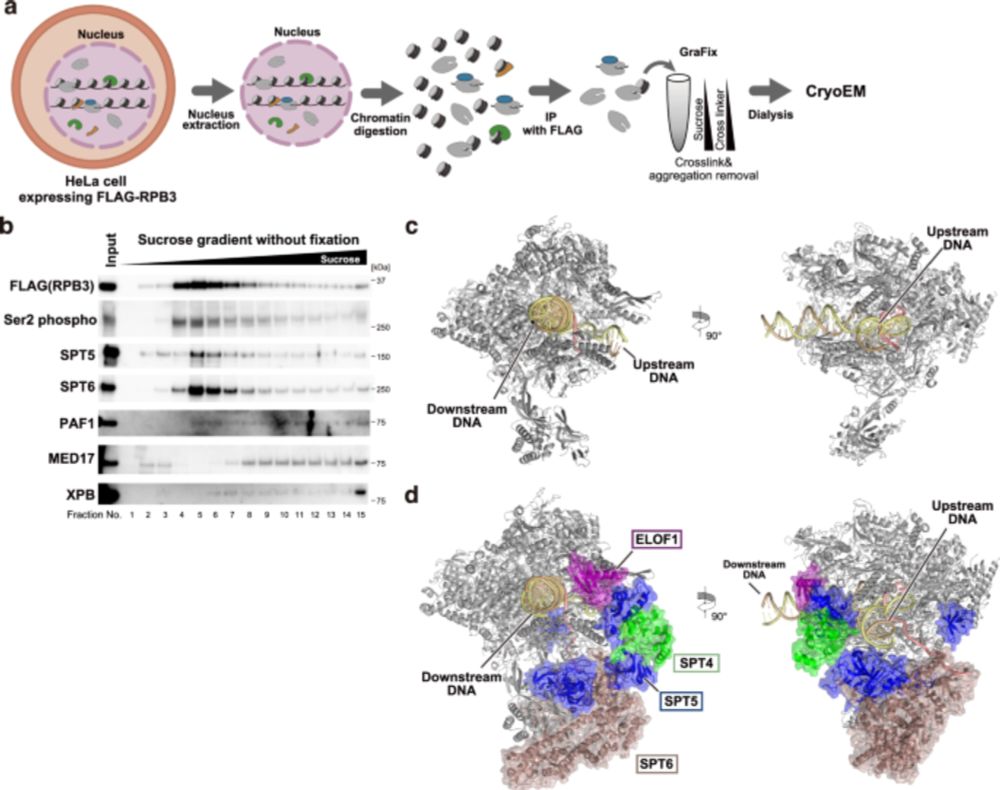
Multiple structures of RNA polymerase II isolated from human nuclei by ChIP-CryoEM analysis - Nature Communications
The authors establish the ChIP-CryoEM method, which combines chromatin immunopurification coupled with cryo-electron microscopy, to visualize native human RNAPII elongation complexes in action, with a...
www.nature.com
Reposted by Ariel Kaplan
Ariel Kaplan
@arielkaplan.bsky.social
· May 27

Intrinsically disordered regions facilitate Msn2 target search to drive promoter selectivity
Transcription factors (TFs) regulate gene expression by binding specific DNA motifs, yet only a fraction of putative sites is occupied in vivo. Intrinsically disordered regions (IDRs) have emerged as ...
www.biorxiv.org
Reposted by Ariel Kaplan
Pierre Bourguet
@p-bourguet.bsky.social
· May 28
Ariel Kaplan
@arielkaplan.bsky.social
· May 27

Intrinsically disordered regions facilitate Msn2 target search to drive promoter selectivity
Transcription factors (TFs) regulate gene expression by binding specific DNA motifs, yet only a fraction of putative sites is occupied in vivo. Intrinsically disordered regions (IDRs) have emerged as ...
www.biorxiv.org
Reposted by Ariel Kaplan
Reuven Gordon
@reuven-gordon.bsky.social
· May 28

Direct observation of conformational dynamics of intrinsically disordered proteins at the single-molecule level
Intrinsically disordered proteins (IDPs) and proteins with intrinsically disordered regions (IDRs) are integral to many biological processes, including neurotransmitter regulation, microtubule regulat...
www.biorxiv.org
Reposted by Ariel Kaplan
Daniel Ibrahim
@danielibrahim.bsky.social
· May 27

Conservation of regulatory elements with highly diverged sequences across large evolutionary distances
Nature Genetics - Combining functional genomic data from mouse and chicken with a synteny-based strategy identifies positionally conserved cis-regulatory elements in the absence of direct sequence...
rdcu.be
Ariel Kaplan
@arielkaplan.bsky.social
· May 28
Ariel Kaplan
@arielkaplan.bsky.social
· May 28
Ariel Kaplan
@arielkaplan.bsky.social
· May 27

Intrinsically disordered regions facilitate Msn2 target search to drive promoter selectivity
Transcription factors (TFs) regulate gene expression by binding specific DNA motifs, yet only a fraction of putative sites is occupied in vivo. Intrinsically disordered regions (IDRs) have emerged as ...
www.biorxiv.org
Ariel Kaplan
@arielkaplan.bsky.social
· May 27
Ariel Kaplan
@arielkaplan.bsky.social
· May 27
Ariel Kaplan
@arielkaplan.bsky.social
· May 27
Ariel Kaplan
@arielkaplan.bsky.social
· May 27