Arriën Symon Rauh
@asrauh.bsky.social
110 followers
180 following
4 posts
PhD Fellow @UCPH computationally studying IDPs in the KLL lab 🇩🇰
Posts
Media
Videos
Starter Packs
Reposted by Arriën Symon Rauh
Reposted by Arriën Symon Rauh
Jules Marien
@marienj.bsky.social
· Sep 3
Reposted by Arriën Symon Rauh
Reposted by Arriën Symon Rauh
Reposted by Arriën Symon Rauh
Arriën Symon Rauh
@asrauh.bsky.social
· Apr 15
Reposted by Arriën Symon Rauh
Sören von Bülow
@sobuelow.bsky.social
· Mar 25
Arriën Symon Rauh
@asrauh.bsky.social
· Mar 21
Reposted by Arriën Symon Rauh
Reposted by Arriën Symon Rauh
Rohit V. Pappu
@rohitpappu68.bsky.social
· Feb 28

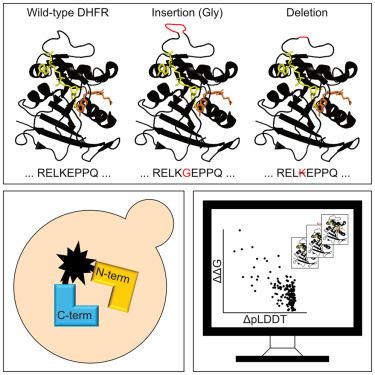
Molecular grammars of intrinsically disordered regions that span the human proteome
Intrinsically disordered regions (IDRs) of proteins are defined by functionally relevant molecular grammars. This refers to IDR-specific non-random amino acid compositions and non-random patterning of...
www.biorxiv.org
Reposted by Arriën Symon Rauh
Reposted by Arriën Symon Rauh
Reposted by Arriën Symon Rauh
Reposted by Arriën Symon Rauh
Reposted by Arriën Symon Rauh
Rohit V. Pappu
@rohitpappu68.bsky.social
· Nov 15
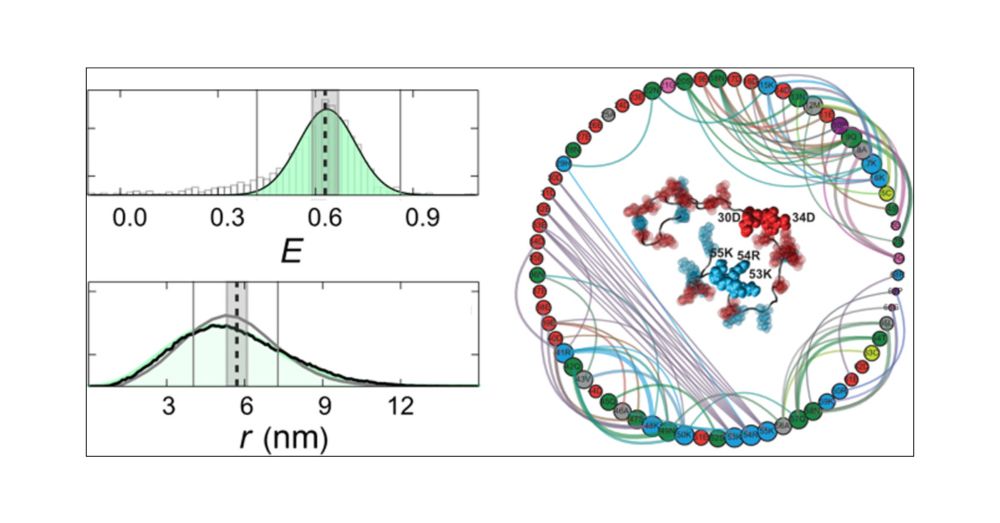
Identifying Sequence Effects on Chain Dimensions of Disordered Proteins by Integrating Experiments and Simulations
It has become increasingly evident that the conformational distributions of intrinsically disordered proteins or regions are strongly dependent on their amino acid compositions and sequence. To facili...
pubs.acs.org