Brian Liau
@brianliau.bsky.social
200 followers
52 following
16 posts
Chemical biologist broadly defined
Posts
Media
Videos
Starter Packs
Brian Liau
@brianliau.bsky.social
· Apr 3

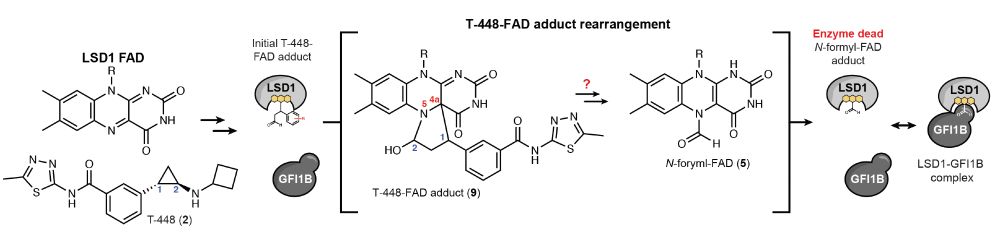
Covalent adduct Grob fragmentation underlies LSD1 demethylase-specific inhibitor mechanism of action and resistance - Nature Communications
Next generation precision lysine-specific histone demethylase 1A (LSD1) covalent inhibitors which selectively block LSD1 enzyme activity by forming a compact N-formyl-FAD adduct have been developed, b...
www.nature.com
Brian Liau
@brianliau.bsky.social
· Mar 27
Brian Liau
@brianliau.bsky.social
· Mar 25
Brian Liau
@brianliau.bsky.social
· Mar 23
Brian Liau
@brianliau.bsky.social
· Feb 27
Brian Liau
@brianliau.bsky.social
· Feb 13
Brian Liau
@brianliau.bsky.social
· Feb 13
Brian Liau
@brianliau.bsky.social
· Feb 13
Brian Liau
@brianliau.bsky.social
· Feb 13
Brian Liau
@brianliau.bsky.social
· Feb 13
Brian Liau
@brianliau.bsky.social
· Feb 12