Calin Plesa
@calin.bsky.social
1.4K followers
930 following
170 posts
Assistant Prof. @uoknightcampus.bsky.social. Cofounder @synplexity.bsky.social. Gene synthesis, synbio, protein engineering, nanopores, multiplex assays. Opinions my own. www.plesalab.org
Posts
Media
Videos
Starter Packs
Pinned
Calin Plesa
@calin.bsky.social
· Jan 28
Reposted by Calin Plesa
Reposted by Calin Plesa
Karl Romanowicz
@kroman.bsky.social
· Aug 16

Exploring antibiotic resistance in diverse homologs of the dihydrofolate reductase protein family through broad mutational scanning
DropSynth technology enables scalable and cost-effective exploration of antibiotic resistance across the DHFR protein family.
www.science.org
Reposted by Calin Plesa
Reposted by Calin Plesa
Reposted by Calin Plesa
Reposted by Calin Plesa
Pascal Notin
@pascalnotin.bsky.social
· May 8
Reposted by Calin Plesa
Calin Plesa
@calin.bsky.social
· Mar 28
Reposted by Calin Plesa
Reposted by Calin Plesa
Calin Plesa
@calin.bsky.social
· Jan 28
Calin Plesa
@calin.bsky.social
· Jan 28
Calin Plesa
@calin.bsky.social
· Jan 28
Calin Plesa
@calin.bsky.social
· Jan 28

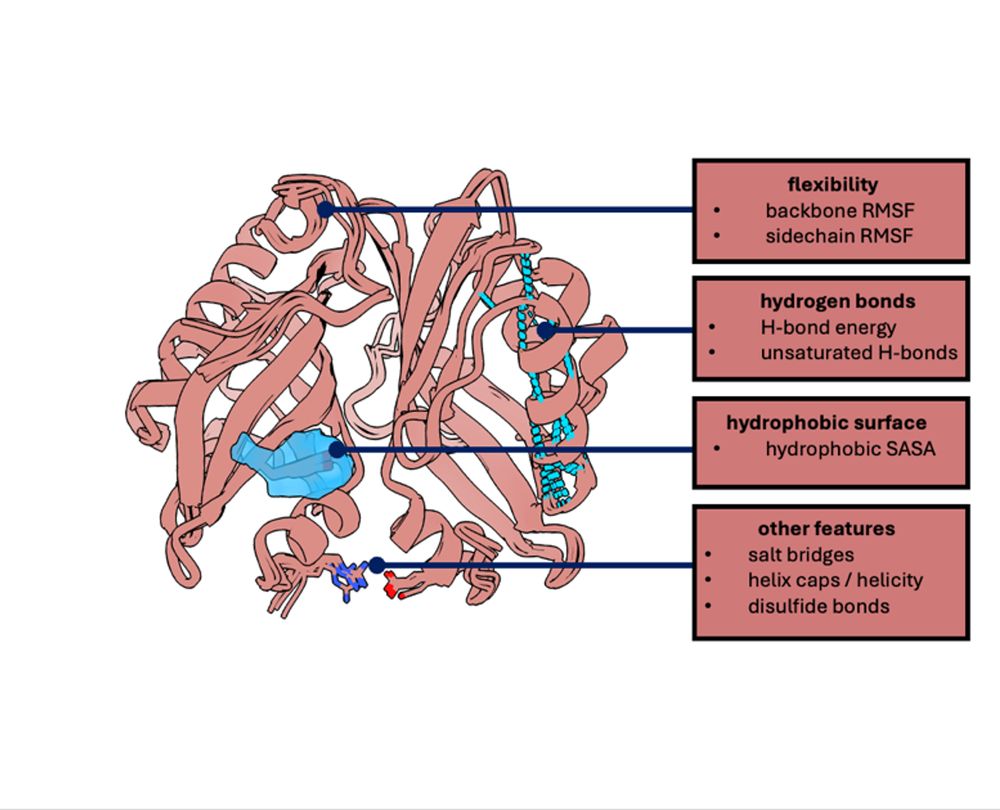
Exploring Antibiotic Resistance in Diverse Homologs of the Dihydrofolate Reductase Protein Family through Broad Mutational Scanning
Current antibiotic resistance studies often focus on individual protein variants, neglecting broader protein family dynamics. Dihydrofolate reductase (DHFR), a key antibiotic target, has been extensiv...
www.biorxiv.org