Carl de Boer
@carldeboer.bsky.social
470 followers
170 following
65 posts
Assistant Professor, UBC school of Biomedical Engineering. Trying to enable personalized medicine by solving gene regulatory code.
Posts
Media
Videos
Starter Packs
Reposted by Carl de Boer
Reposted by Carl de Boer
John Ray
@jraylab.bsky.social
· 19d

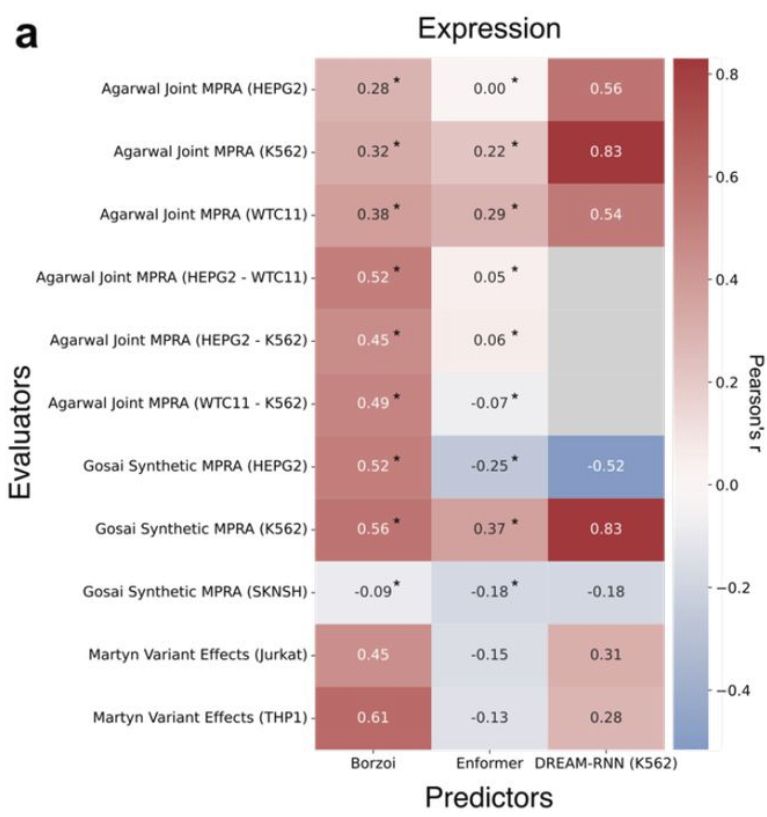
Genetic and epigenetic screens in primary human T cells link candidate causal autoimmune variants to T cell networks - Nature Genetics
Massively parallel reporter assay in primary human CD4+ T cells and bulk and single-cell CRISPR-interference screens identify candidate causal variants linked to autoimmune disease risk that modulate ...
www.nature.com
Carl de Boer
@carldeboer.bsky.social
· Sep 4
Reposted by Carl de Boer
Cyntia Taveneau
@cyntiat.bsky.social
· Sep 2

Code to complex: AI-driven de novo binder design
In this review, Fox et al. discuss how artificial intelligence has transformed our
ability to design new-to-nature proteins that bind target proteins with high affinity
and specificity. The authors de...
www.cell.com
Reposted by Carl de Boer
Shaun Mahony
@shaunmahony.bsky.social
· Aug 23

Optimized ChIP-exo for mammalian cells and patterned sequencing flow cells
By combining chromatin immunoprecipitation (ChIP) with an exonuclease digestion of protein-bound DNA fragments, ChIP-exo characterizes genome-wide protein-DNA interactions at near base-pair resolution...
www.biorxiv.org
Reposted by Carl de Boer
Matthew Aguirre
@aguirre404.bsky.social
· Aug 22
Reposted by Carl de Boer
Reposted by Carl de Boer
Carl de Boer
@carldeboer.bsky.social
· Jul 11
Carl de Boer
@carldeboer.bsky.social
· Jul 11
Carl de Boer
@carldeboer.bsky.social
· Jul 11
Carl de Boer
@carldeboer.bsky.social
· Jul 11
Carl de Boer
@carldeboer.bsky.social
· Jul 11