Tom Ellis
@proftomellis.bsky.social
3.1K followers
350 following
290 posts
Synthetic Biology & Synthetic Genomics @ Imperial College London and the Sanger Institute. Bilingual in English and DNA. Views are either my own or my microbes'
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Tom Ellis
Tom Ellis
@proftomellis.bsky.social
· Aug 19
Tom Ellis
@proftomellis.bsky.social
· Aug 19
Tom Ellis
@proftomellis.bsky.social
· Aug 19
Tom Ellis
@proftomellis.bsky.social
· Aug 19

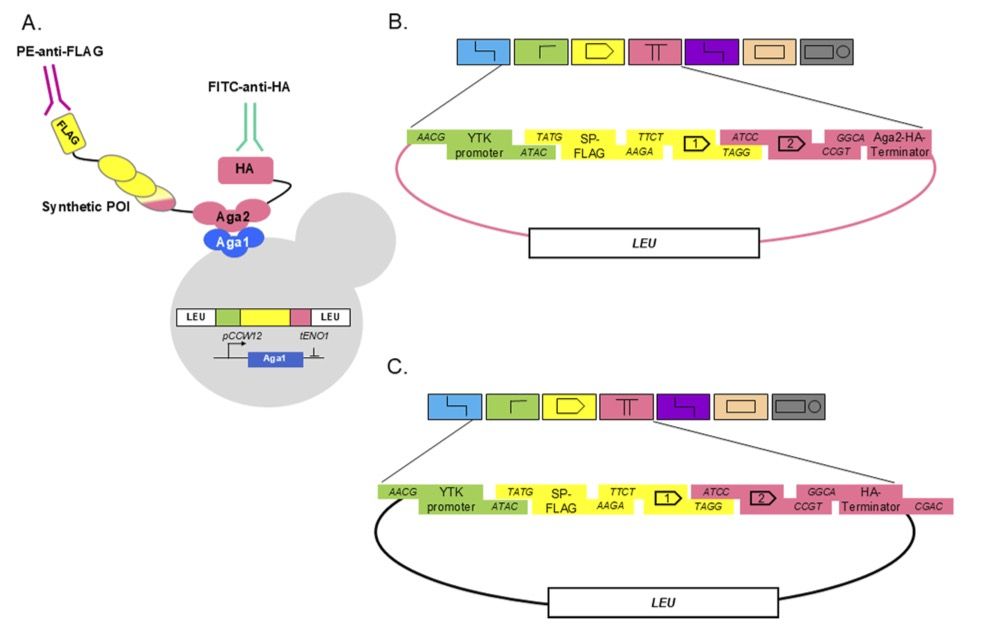
Combinatorial Design Testing in Genomes with POLAR-seq
Synthetic biology projects increasingly use modular DNA assembly or synthetic in vivo recombination to generate diverse combinatorial libraries of genetic constructs for testing. But as these designs ...
www.biorxiv.org
Tom Ellis
@proftomellis.bsky.social
· Aug 19
Tom Ellis
@proftomellis.bsky.social
· Aug 19
Reposted by Tom Ellis
Tom Ellis
@proftomellis.bsky.social
· Aug 13
Reposted by Tom Ellis
Tom Ellis
@proftomellis.bsky.social
· Aug 11
Tom Ellis
@proftomellis.bsky.social
· Aug 11