Robert E. Campbell
@campbell-lab.bsky.social
1.7K followers
2.6K following
16 posts
Professor in the Department of Chemistry, School of Science, The University of Tokyo.
Editor-in-Chief of Protein Engineering, Design & Selection (PEDS).
Opinions are my own.
https://campbell.chem.s.u-tokyo.ac.jp
Posts
Media
Videos
Starter Packs
Reposted by Robert E. Campbell
Abhi Aggarwal
@abhiaggarwal.bsky.social
· Jul 31

A sensitive orange fluorescent calcium ion indicator for imaging neural activity
Genetically encoded calcium indicators (GECIs) are vital tools for fluorescence-based visualization of neuronal activity with high spatial and temporal resolution. However, current highest-performance...
www.biorxiv.org
Reposted by Robert E. Campbell
Shinya Tsukiji
@shinyatsukiji.bsky.social
· Jul 26
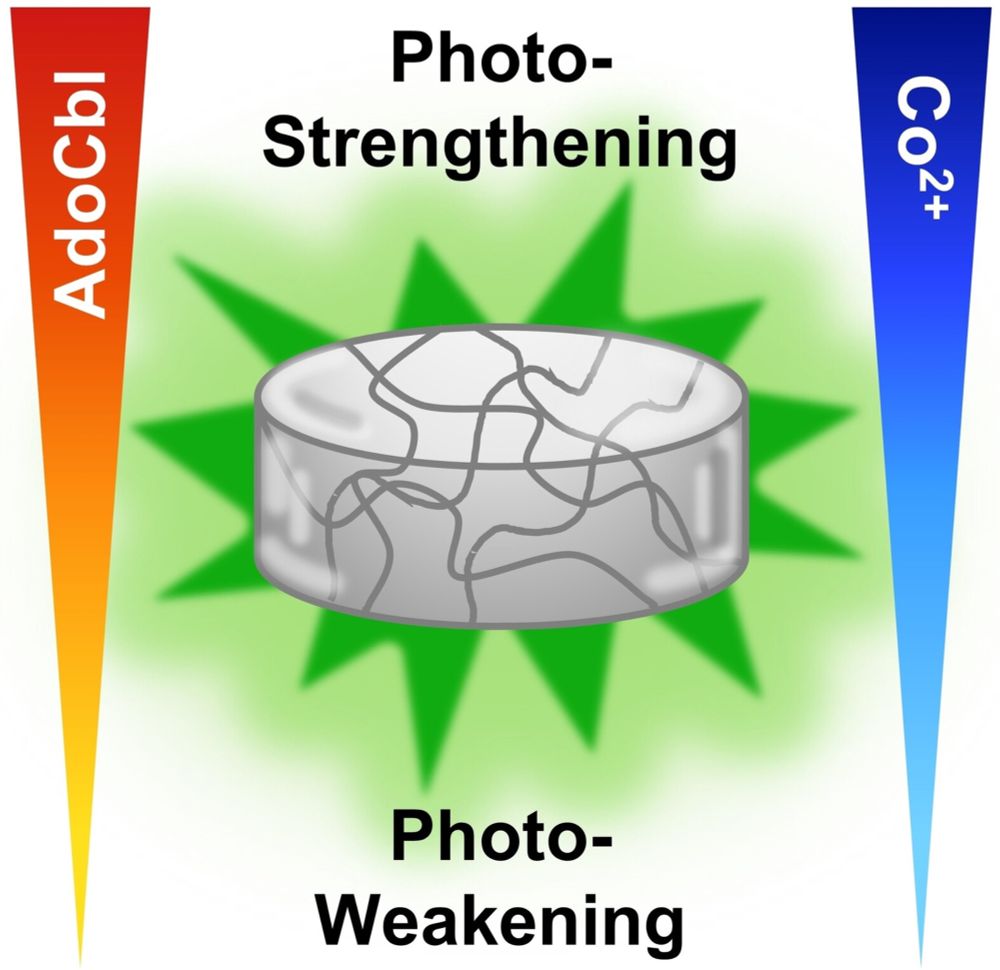
Programming Green Light‐Responsive Hydrogels: From Photo‐Weakening to Photo‐Unresponsiveness and Photo‐Strengthening
The light response of CarHc protein-based hydrogels can be alterted from photo-weakening, to photo-strengthening, or becoming light-unresponsive depending on the added cofactors AdoCbl and Co2+. Unde....
chemistry-europe.onlinelibrary.wiley.com
Reposted by Robert E. Campbell
Reposted by Robert E. Campbell