Chris de Graaf
@cdg-gpcr.bsky.social
290 followers
310 following
21 posts
Head of Computational Drug Discovery and Data Science at Structure Therapeutics | GPCR | SBDD | CADD | Cheminformatics | Computational Chemistry | Medicinal Chemistry | Drug Design
Posts
Media
Videos
Starter Packs
Reposted by Chris de Graaf
Chris de Graaf
@cdg-gpcr.bsky.social
· Jul 2
Identification of nanomolar adenosine A2A receptor ligands using reinforcement learning and structure-based drug design - Nature Communications
Here the authors combine a deep generative model with structure-based drug design and prospectively validate functionally active, nanomolar, A2A adenosine receptor ligands and solve their crystal stru...
www.nature.com
Chris de Graaf
@cdg-gpcr.bsky.social
· Jul 2

Identification of nanomolar adenosine A2A receptor ligands using reinforcement learning and structure-based drug design - Nature Communications
Here the authors combine a deep generative model with structure-based drug design and prospectively validate functionally active, nanomolar, A2A adenosine receptor ligands and solve their crystal stru...
www.nature.com
Reposted by Chris de Graaf
Reposted by Chris de Graaf
Reposted by Chris de Graaf
Chris de Graaf
@cdg-gpcr.bsky.social
· Jan 10
Chris de Graaf
@cdg-gpcr.bsky.social
· Jan 10

Structural and functional determination of peptide versus small molecule ligand binding at the apelin receptor - Nature Communications
This study explores apelin receptor’s role in cardiovascular function, identifying residues critical for binding through genetic variants, AlphaFold2 modelling and base editing in cardiomyocytes. Co-c...
www.nature.com
Reposted by Chris de Graaf
Chris de Graaf
@cdg-gpcr.bsky.social
· Dec 15
Chris de Graaf
@cdg-gpcr.bsky.social
· Dec 15
Reposted by Chris de Graaf
Chris de Graaf
@cdg-gpcr.bsky.social
· Dec 9
More response to the response (DiffDock/DiffDock-L)
To amplify on what Pat Walters said (quotes from Gabriel Corso's post in italics): It only took a few days (in August 2023) for Ann Cleves, Pat, and me to figure out that the original DiffDock work ha...
www.linkedin.com
Reposted by Chris de Graaf
Chris de Graaf
@cdg-gpcr.bsky.social
· Dec 1
Reposted by Chris de Graaf
Reposted by Chris de Graaf
Chris de Graaf
@cdg-gpcr.bsky.social
· Nov 23
Chris de Graaf
@cdg-gpcr.bsky.social
· Nov 23

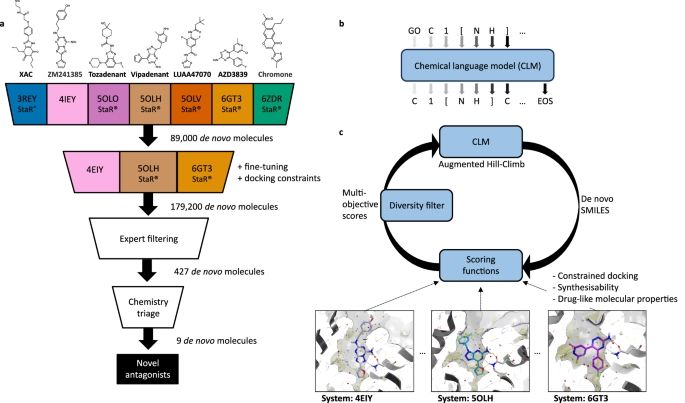
Modern hit-finding with structure-guided de novo design: identification of novel nanomolar adenosine A2A receptor ligands using reinforcement learning
Generative chemical language models have demonstrated success in learning language-based molecular representations for de novo drug design. Here, we integrate structure-based drug design (SBDD) princi...
chemrxiv.org
Reposted by Chris de Graaf
Chris de Graaf
@cdg-gpcr.bsky.social
· Nov 23

Comparative Study of Allosteric GPCR Binding Sites and Their Ligandability Potential
The steadily growing number of experimental G-protein-coupled receptor (GPCR) structures has revealed diverse locations of allosteric modulation, and yet few drugs target them. This gap highlights the...
pubs.acs.org
Reposted by Chris de Graaf
Chris de Graaf
@cdg-gpcr.bsky.social
· Nov 23

Modern hit-finding with structure-guided de novo design: identification of novel nanomolar adenosine A2A receptor ligands using reinforcement learning
Generative chemical language models have demonstrated success in learning language-based molecular representations for de novo drug design. Here, we integrate structure-based drug design (SBDD) princi...
chemrxiv.org
Reposted by Chris de Graaf
Chris de Graaf
@cdg-gpcr.bsky.social
· Dec 1
Chris de Graaf
@cdg-gpcr.bsky.social
· Dec 1
Reposted by Chris de Graaf
Reposted by Chris de Graaf
Chris de Graaf
@cdg-gpcr.bsky.social
· Nov 28