Posts
Media
Videos
Starter Packs
Pinned
Chris Fell
@cfell.bsky.social
· Apr 9
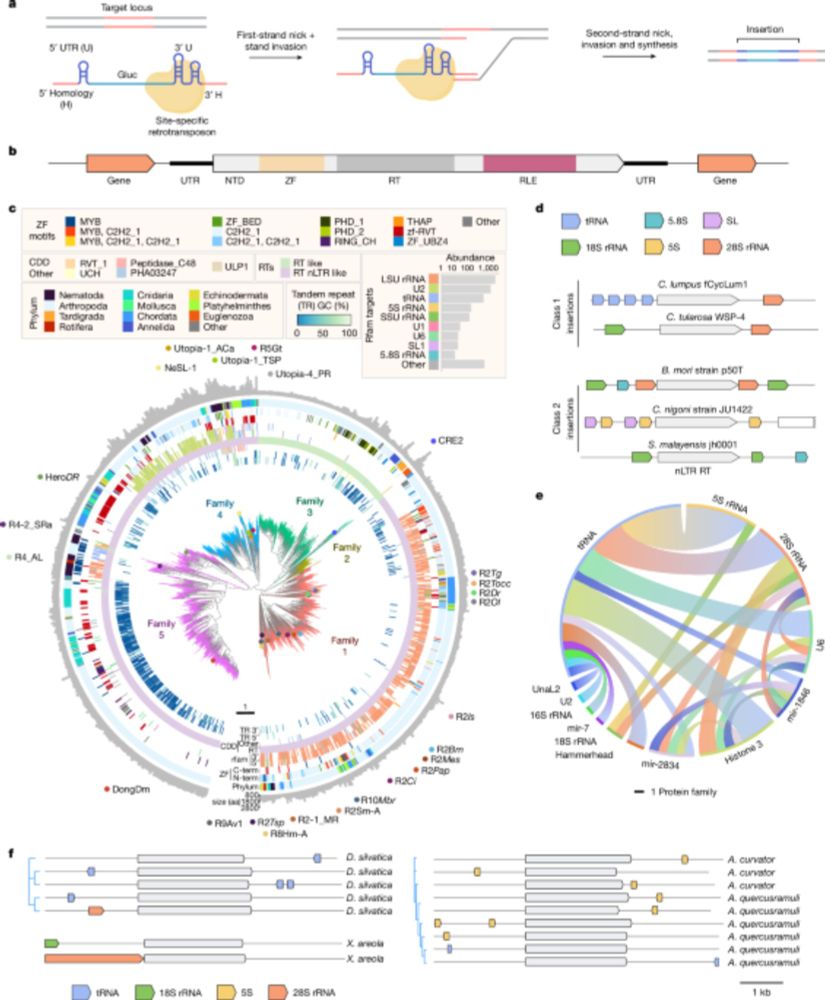
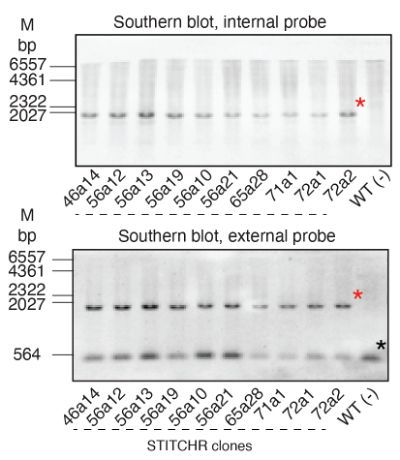
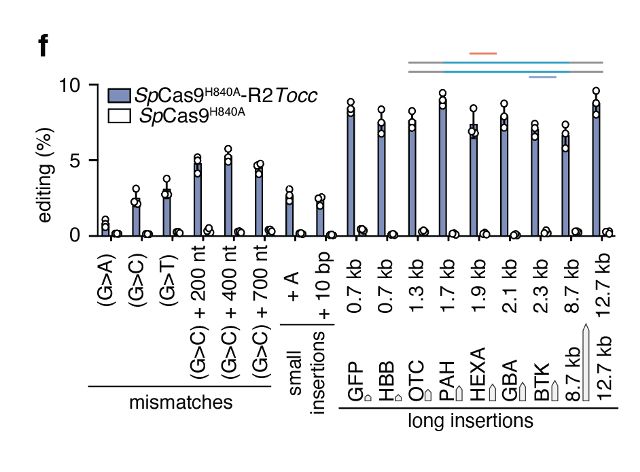
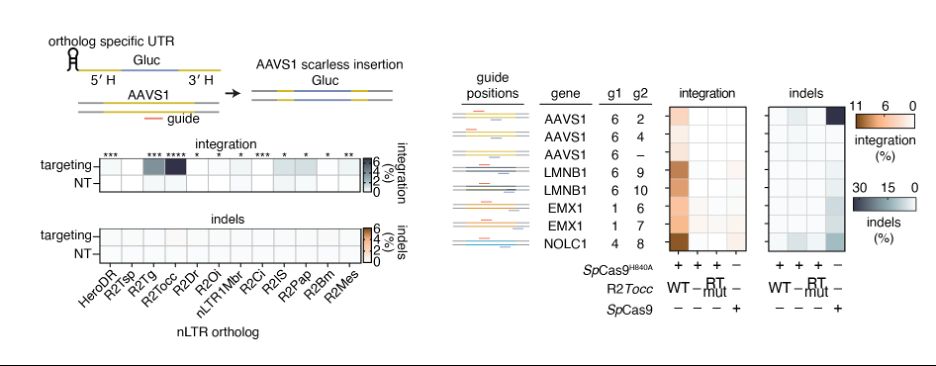
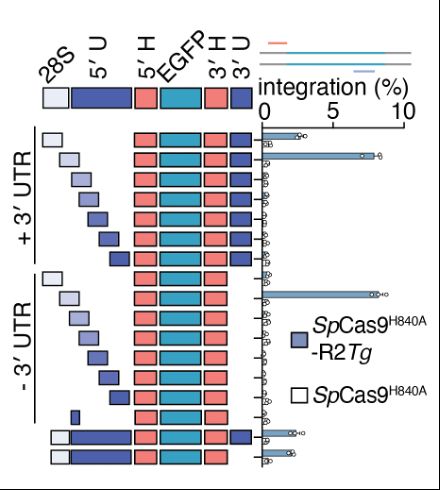
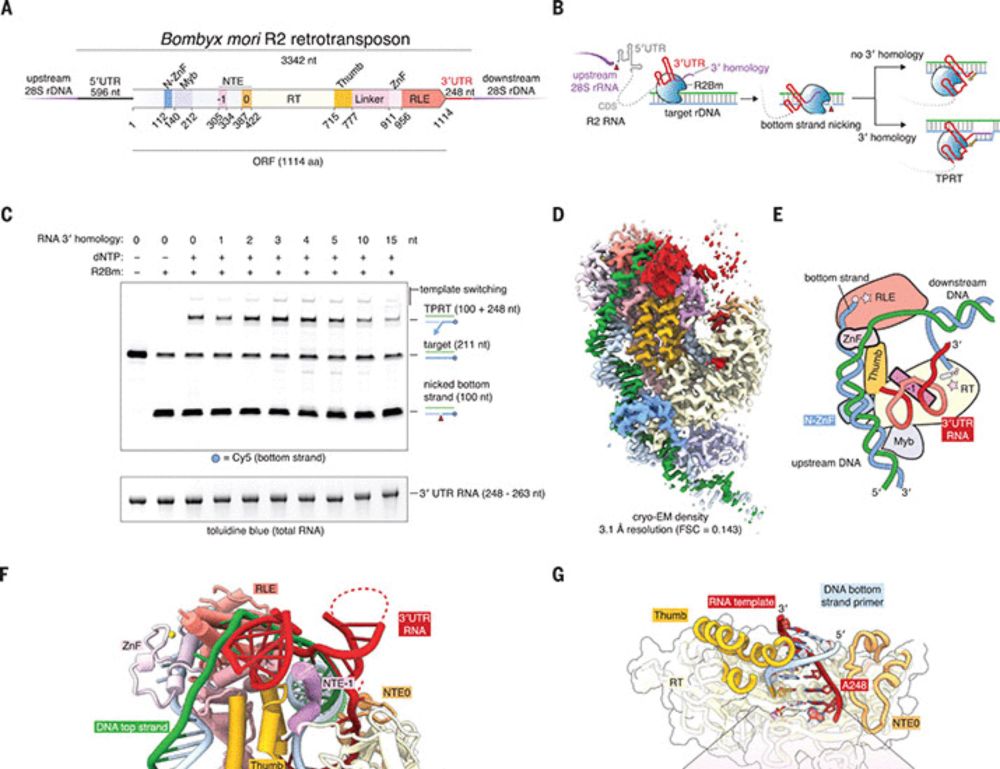
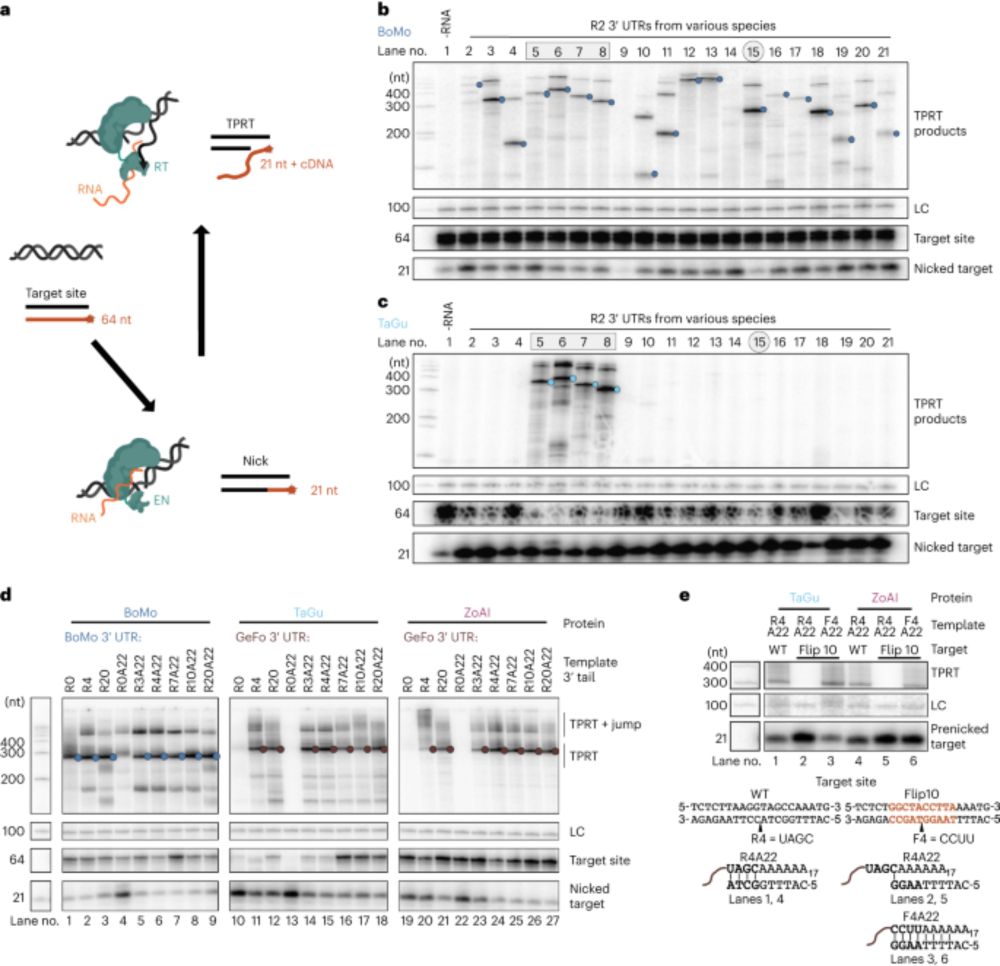
Reprogramming site-specific retrotransposon activity to new DNA sites - Nature
A study of retrotransposon activity repurposes a retroelement called R2Tocc to create a programmable system called STITCHR that enables diverse genome edits including efficient, scarless large pa...
www.nature.com
Reposted by Chris Fell
Reposted by Chris Fell
Addgene
@addgene.bsky.social
· Apr 10
Chris Fell
@cfell.bsky.social
· Apr 10
Chris Fell
@cfell.bsky.social
· Apr 9
Chris Fell
@cfell.bsky.social
· Apr 9
Chris Fell
@cfell.bsky.social
· Apr 9
Chris Fell
@cfell.bsky.social
· Apr 9

Reprogramming site-specific retrotransposon activity to new DNA sites - Nature
A study of retrotransposon activity repurposes a retroelement called R2Tocc to create a programmable system called STITCHR that enables diverse genome edits including efficient, scarless large pa...
www.nature.com
Chris Fell
@cfell.bsky.social
· Dec 16
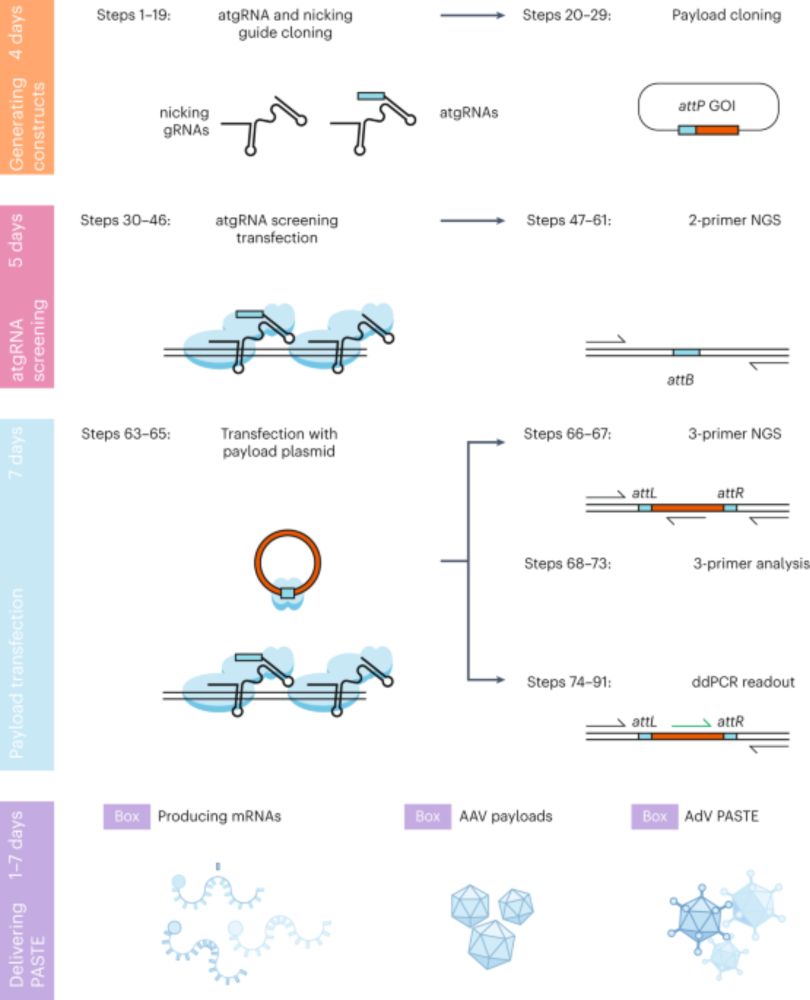
Precise kilobase-scale genomic insertions in mammalian cells using PASTE - Nature Protocols
Programmable addition via site-specific targeting elements (PASTE) combines the specificity, efficiency and cargo size benefits of site-specific integrases with the programmability of prime editing fo...
www.nature.com