Between Biology, Data & Care 🔬🧬
Sarcomas & rare cancers | Affiliated with Gustave Roussy

🔓 rdcu.be/eQmQp

🔓 rdcu.be/eQmQp


🔗 www.biorxiv.org/content/10.1...

🔗 www.biorxiv.org/content/10.1...
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
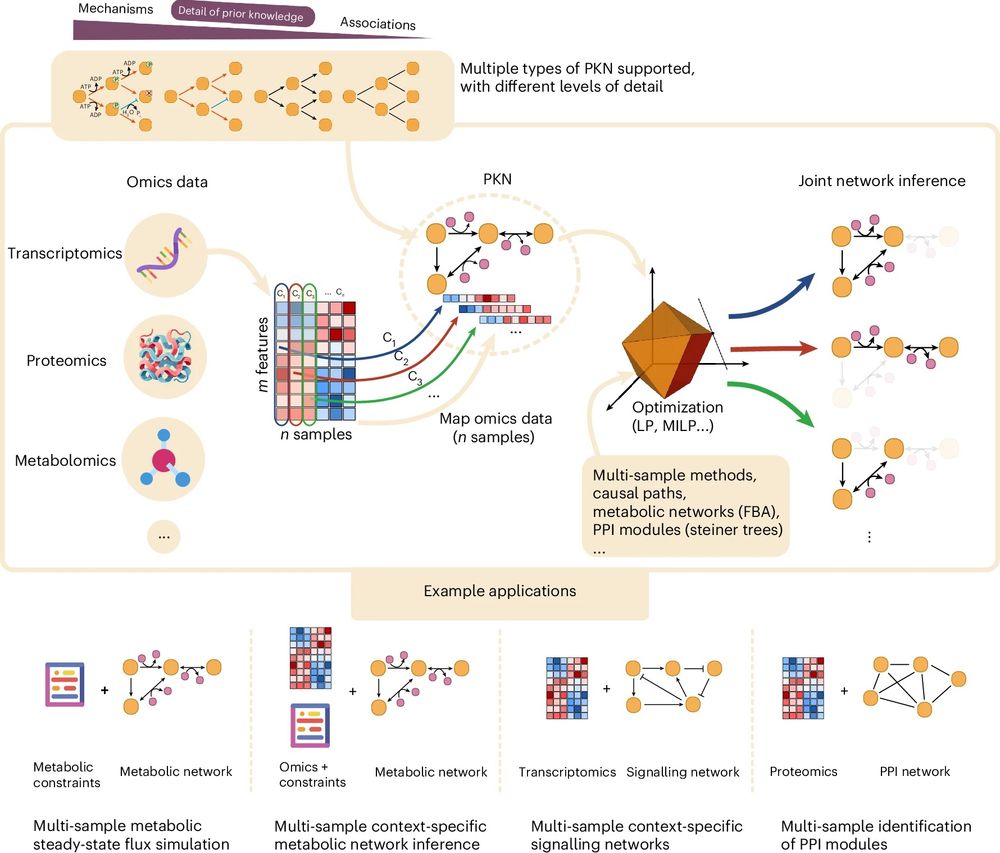
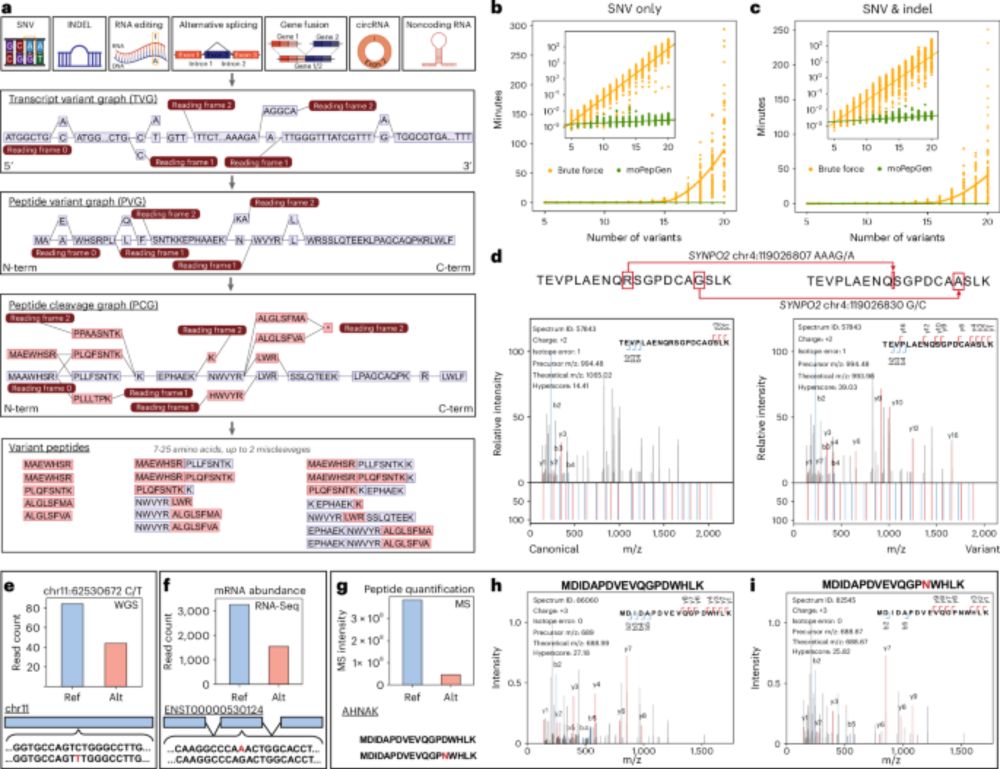
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇


Introducing C2S‑Scale—a Yale and Google collab: we scaled LLMs (up to 27B!) to analyze & generate single‑cell data 🧬 ➡️ 📝
🔗 Blog: research.google/blog/teachin...
🔗 Preprint: biorxiv.org/content/10.1...

Introducing C2S‑Scale—a Yale and Google collab: we scaled LLMs (up to 27B!) to analyze & generate single‑cell data 🧬 ➡️ 📝
🔗 Blog: research.google/blog/teachin...
🔗 Preprint: biorxiv.org/content/10.1...
www.nature.com/artic...
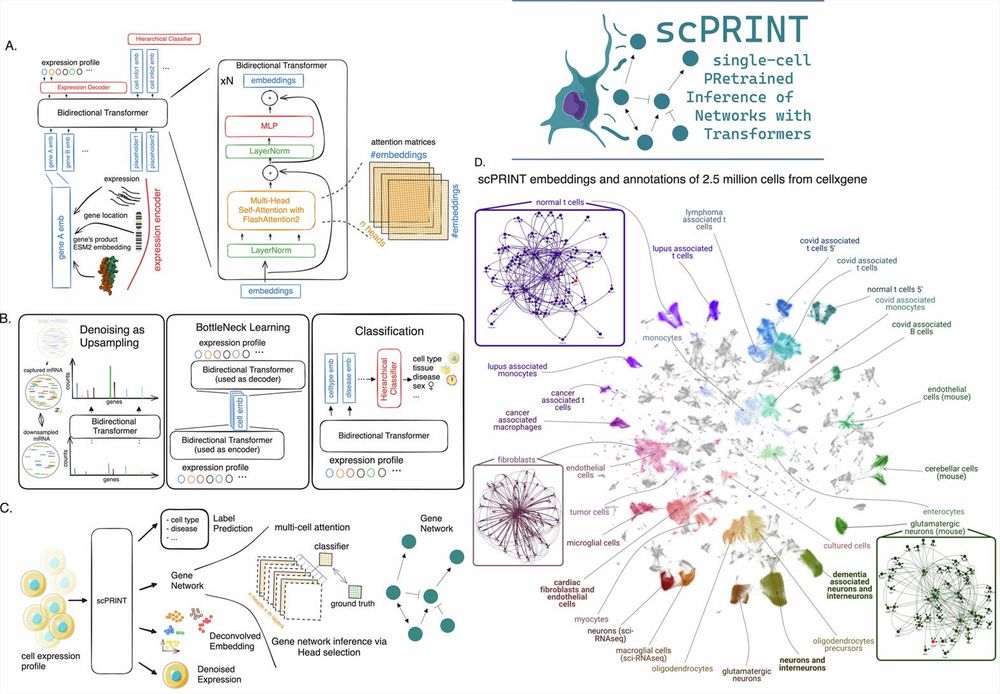
www.nature.com/artic...

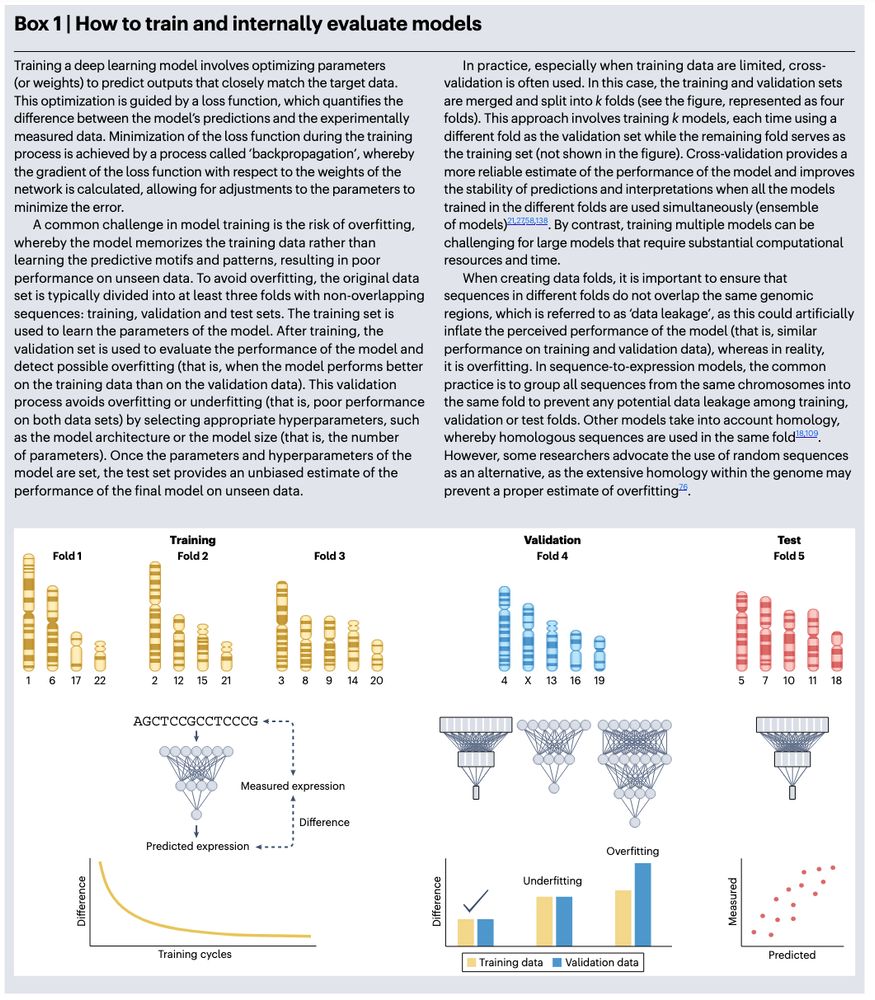
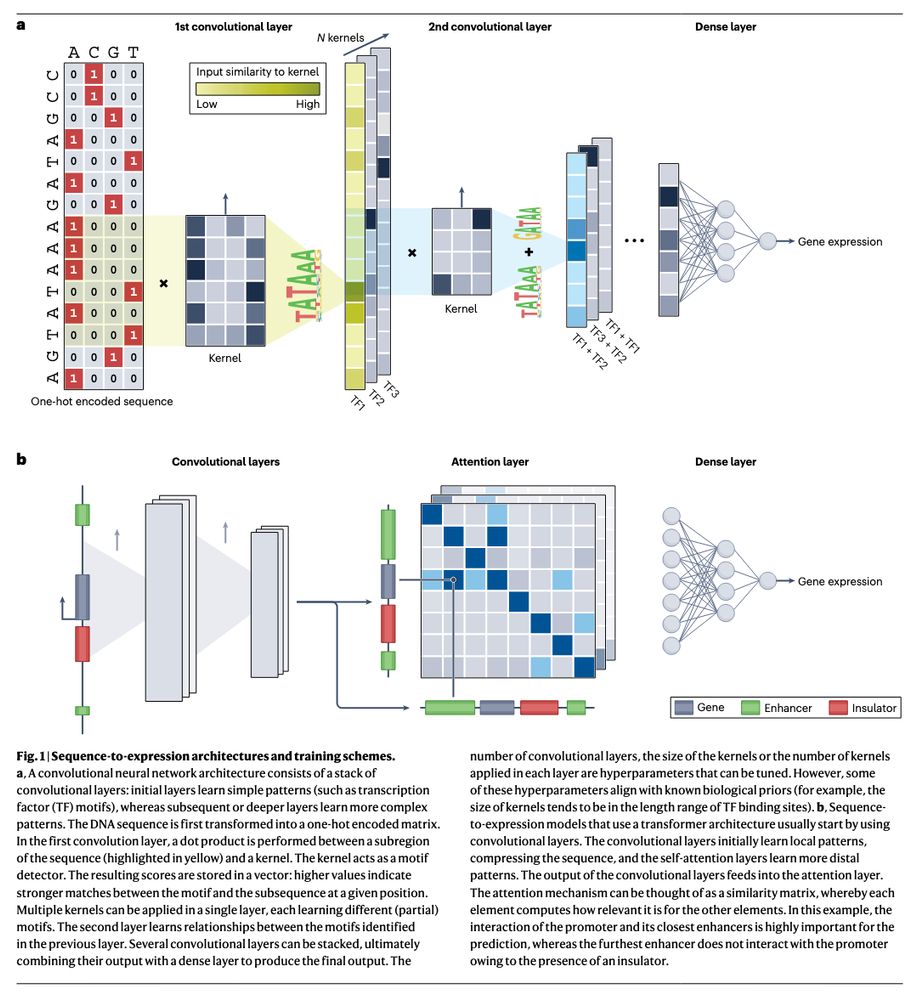
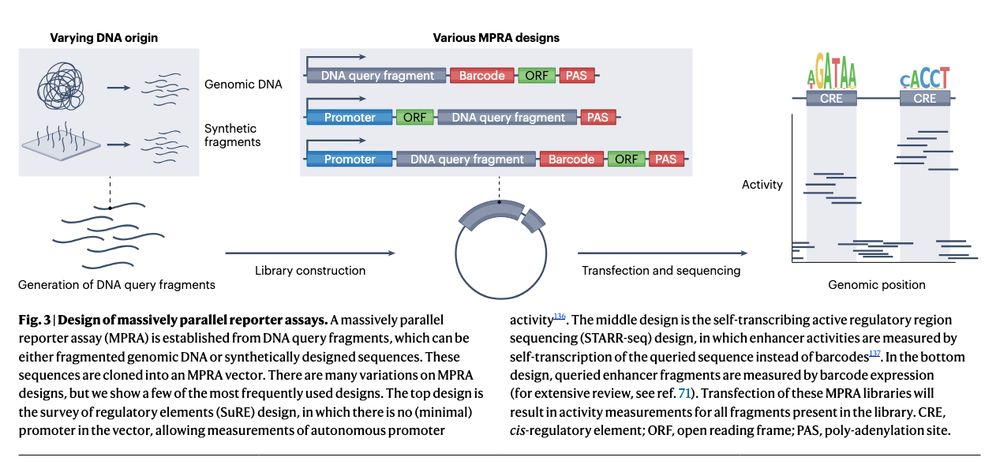
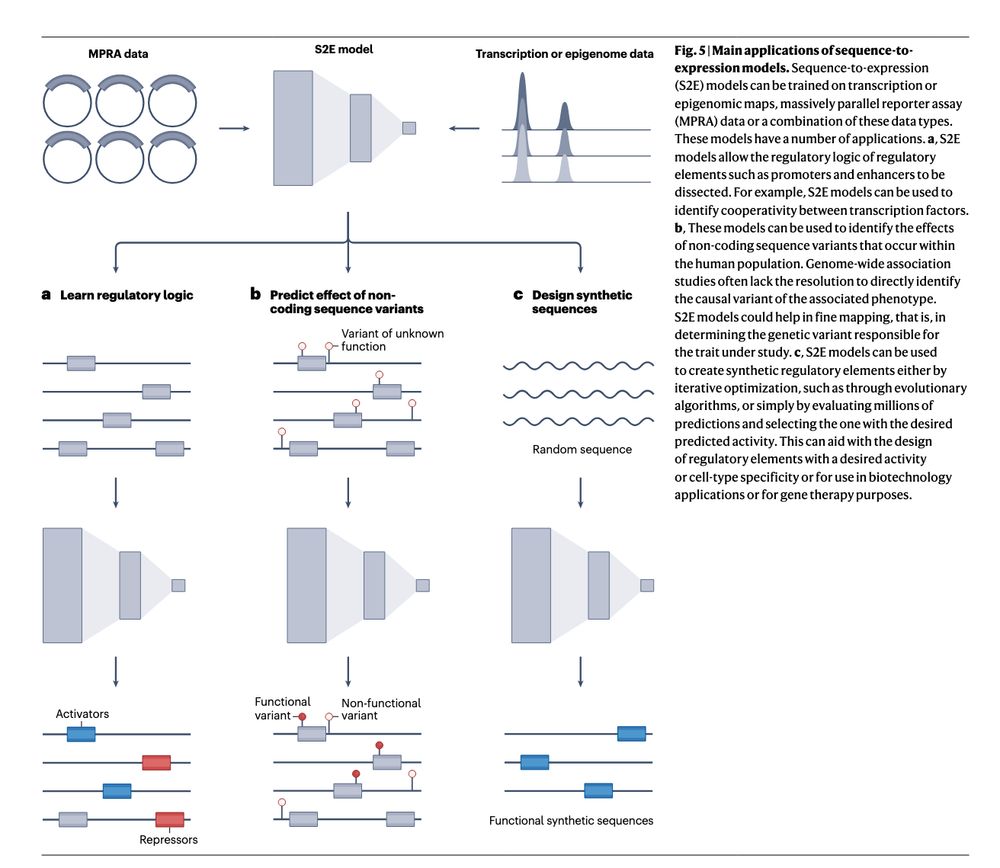
https://go.nature.com/3YpvALv

https://go.nature.com/3YpvALv
https://go.nature.com/3YvGcbJ

https://go.nature.com/3YvGcbJ

For this reason, Elyas Heidari, a student in my lab and in @steglelab.bsky.social built segger.
www.biorxiv.org/content/10.1...
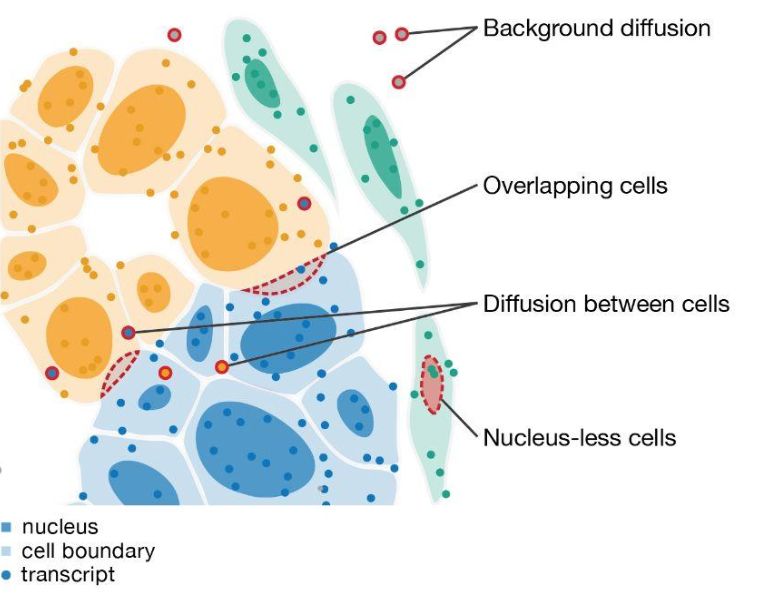
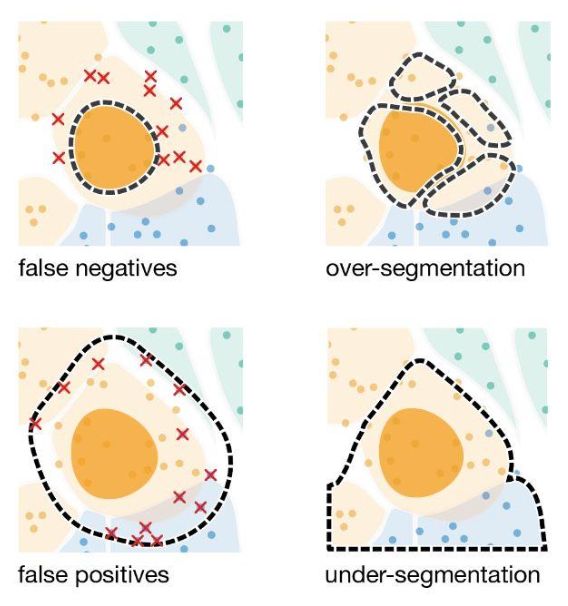
For this reason, Elyas Heidari, a student in my lab and in @steglelab.bsky.social built segger.
www.biorxiv.org/content/10.1...
Comprehensive #DSRCT multi-omics analyses unveil CACNA2D2 as a diagnostic hallmark and super-enhancer-driven EWSR1::WT1 signature gene
tinyurl.com/42fxmmud
@dkfz.bsky.social
@uniheidelberg.bsky.social
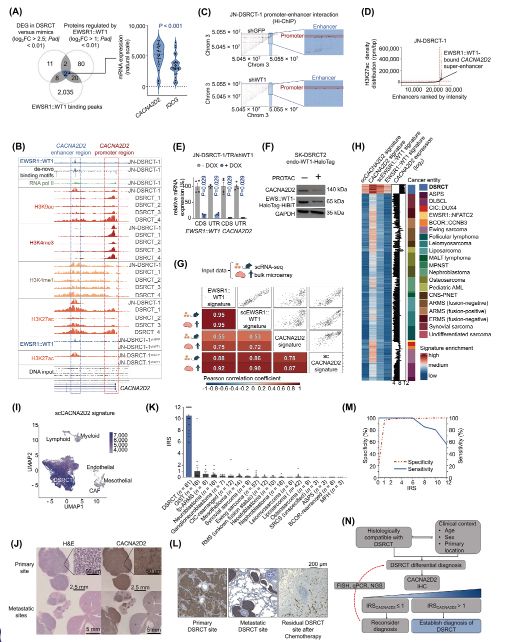
Comprehensive #DSRCT multi-omics analyses unveil CACNA2D2 as a diagnostic hallmark and super-enhancer-driven EWSR1::WT1 signature gene
tinyurl.com/42fxmmud
@dkfz.bsky.social
@uniheidelberg.bsky.social
Wichtiger, denn je!
www.openpetition.de/petition/onl...
Hier noch Bilder aus #Essen UK-Essen und #Fortress Meeting. #Aufstehenfuerdemokratie


Wichtiger, denn je!
www.openpetition.de/petition/onl...
Hier noch Bilder aus #Essen UK-Essen und #Fortress Meeting. #Aufstehenfuerdemokratie


