Gautam Shirsekar
@coevolution.bsky.social
76 followers
64 following
24 posts
Plant pathologist | Genomics | Coevolution | Wild plant pathosystems | Weigelworld, Max Planck Institute for Biology | Asst. Prof. at the University of Tennessee, Knoxville (EPP)| www.coevolutionlab.org
Posts
Media
Videos
Starter Packs
Pinned
Gautam Shirsekar
@coevolution.bsky.social
· Aug 14
Reposted by Gautam Shirsekar
Gautam Shirsekar
@coevolution.bsky.social
· Aug 15
Gautam Shirsekar
@coevolution.bsky.social
· Aug 14
Gautam Shirsekar
@coevolution.bsky.social
· Aug 14
Gautam Shirsekar
@coevolution.bsky.social
· Aug 14
Reposted by Gautam Shirsekar
K.D. Murray
@kdm9.bsky.social
· Aug 14
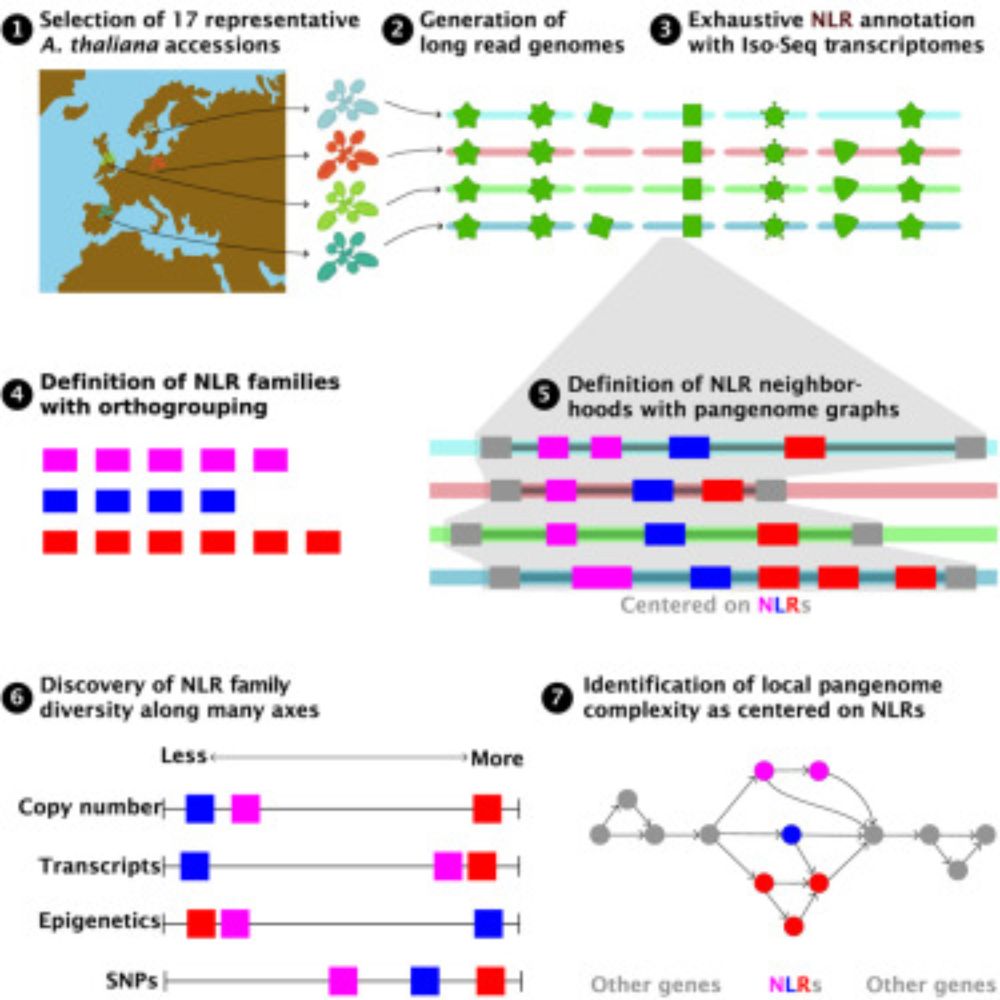
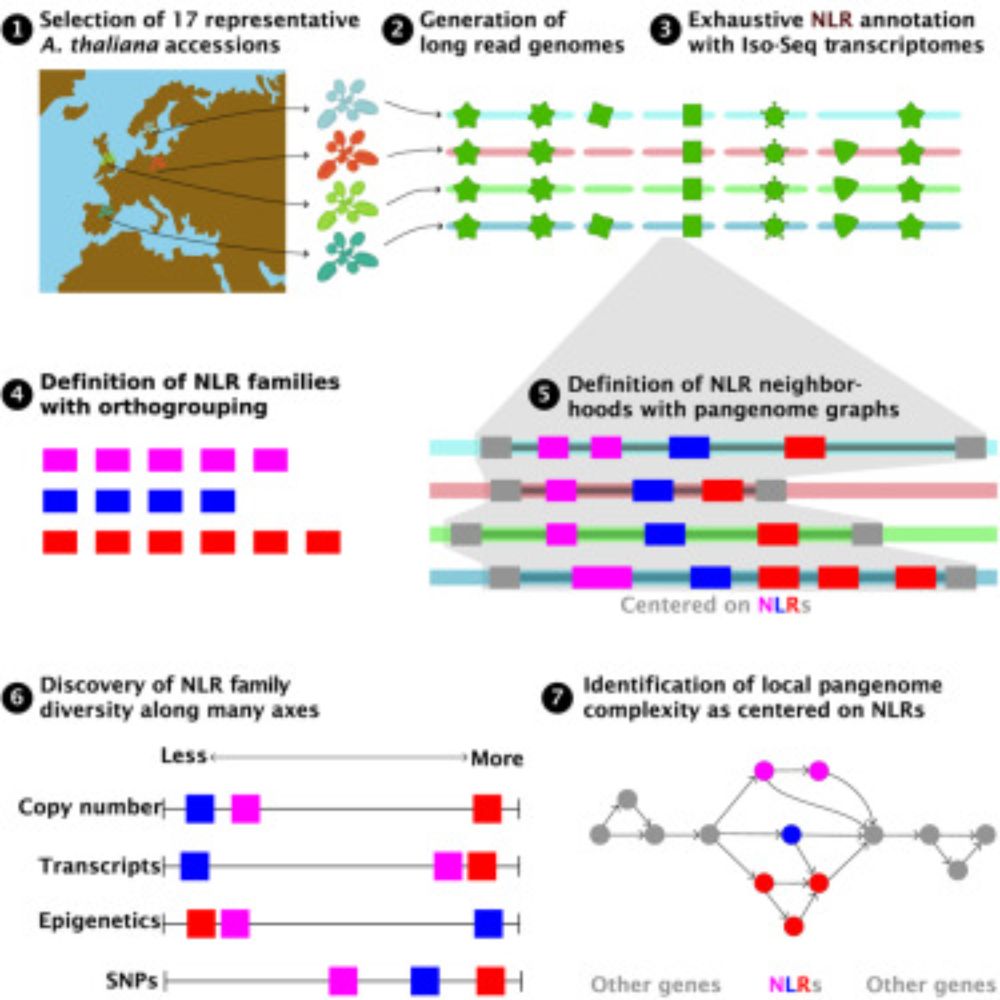
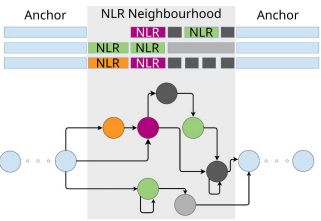
Pangenomic context reveals the extent of intraspecific plant NLR evolution
Individual- and population-level diversity is required for pathogen defense by nucleotide-binding
site leucine-rich repeat (NLR) proteins. Teasdale et al. leverage annotated, divergent
A. thaliana gen...
www.cell.com
Reposted by Gautam Shirsekar
Reposted by Gautam Shirsekar
K.D. Murray
@kdm9.bsky.social
· Aug 14
Reposted by Gautam Shirsekar
Reposted by Gautam Shirsekar
Gautam Shirsekar
@coevolution.bsky.social
· Aug 14
K.D. Murray
@kdm9.bsky.social
· Aug 14

Pangenomic context reveals the extent of intraspecific plant NLR evolution
Individual- and population-level diversity is required for pathogen defense by nucleotide-binding
site leucine-rich repeat (NLR) proteins. Teasdale et al. leverage annotated, divergent
A. thaliana gen...
www.cell.com