PDRA at LAMMASU-UFCSPA
We present eRMSF, a Python package for ensemble-based RMSF analysis of biomolecular systems, supporting MD, PDB, and ML ensembles.
Proud to collaborate with Rodrigo Ligabue-Braun and @conradopedebos.bsky.social
pubs.acs.org/doi/10.1021/...

We present eRMSF, a Python package for ensemble-based RMSF analysis of biomolecular systems, supporting MD, PDB, and ML ensembles.
Proud to collaborate with Rodrigo Ligabue-Braun and @conradopedebos.bsky.social
pubs.acs.org/doi/10.1021/...
First, we made MD simulations rain down from the cloud. Now, we’re bringing deep learning into the mix! 🌩️🤖

First, we made MD simulations rain down from the cloud. Now, we’re bringing deep learning into the mix! 🌩️🤖
➡️ AlphaBridge will identify the most confident interactions, making your analysis easier!
🌐Try it here: alpha-bridge.eu
👀Read more: www.biorxiv.org/content/10.1...

➡️ AlphaBridge will identify the most confident interactions, making your analysis easier!
🌐Try it here: alpha-bridge.eu
👀Read more: www.biorxiv.org/content/10.1...
Here’s our latest work on parametrising coarse-grained models of sugars (disaccharides) by Astrid Brandner & in collaboration with Iain Smith, @pauloctsouza.bsky.social and @cg-martini.bsky.social pubs.acs.org/doi/10.1021/... #glycotime
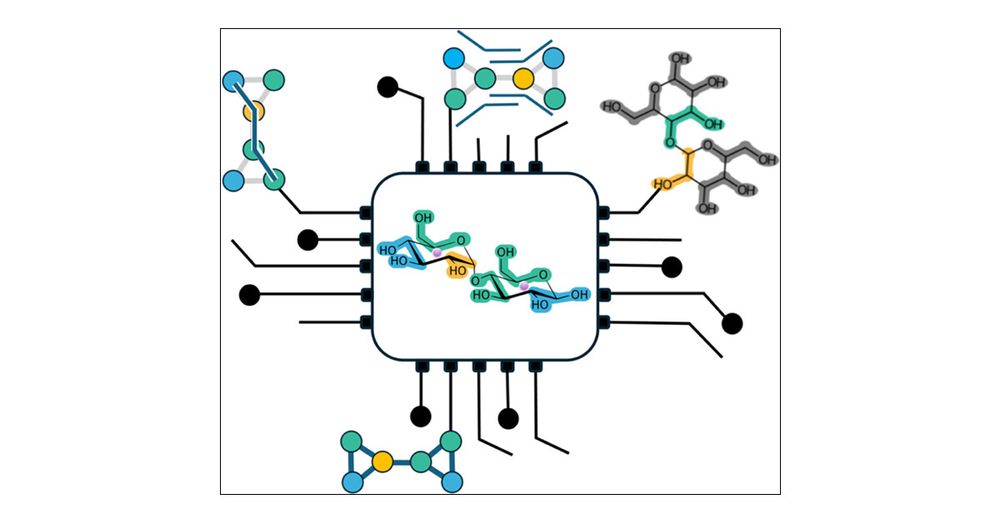
Here’s our latest work on parametrising coarse-grained models of sugars (disaccharides) by Astrid Brandner & in collaboration with Iain Smith, @pauloctsouza.bsky.social and @cg-martini.bsky.social pubs.acs.org/doi/10.1021/... #glycotime
Out now in Angew Chemie
onlinelibrary.wiley.com/doi/10.1002/...

Out now in Angew Chemie
onlinelibrary.wiley.com/doi/10.1002/...

colab.research.google.com/drive/1GGJKr...
Paper: t.co/evNxaX2omW

colab.research.google.com/drive/1GGJKr...
Paper: t.co/evNxaX2omW
@jbiolchem
Polymyxin B1 in the E. coli inner membrane: a complex story of protein and lipopolysaccharide mediated insertion

