Daniel Petras
@daniel-petras.bsky.social
230 followers
200 following
18 posts
Dad, Punk Rocker, Scientist, Ocean Lover.
Working with the Functional Metabolomics Lab on developing mass spec tools to understand microbial communities.
www.functional-metabolomics.com
Posts
Media
Videos
Starter Packs
Daniel Petras
@daniel-petras.bsky.social
· Jul 29
Daniel Petras
@daniel-petras.bsky.social
· Jul 27
Daniel Petras
@daniel-petras.bsky.social
· Jul 27
Daniel Petras
@daniel-petras.bsky.social
· Jul 27
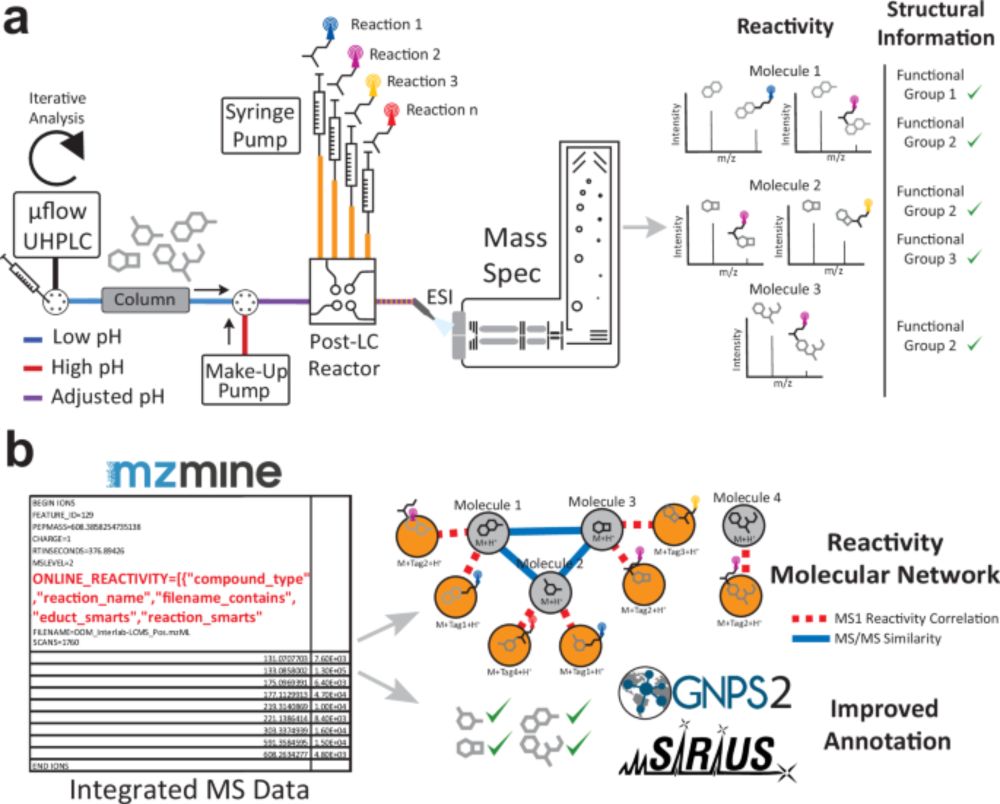
Enhancing tandem mass spectrometry-based metabolite annotation with online chemical labeling - Nature Communications
To improve annotation in non-targeted metabolomics studies, authors develop a Multiplexed Chemical Metabolomics (MCheM) platform, combining post-column derivatization with integrated data processing. ...
www.nature.com
Reposted by Daniel Petras
Reposted by Daniel Petras
Reposted by Daniel Petras
Reposted by Daniel Petras
Mingxun Wang
@mingxunwang.bsky.social
· May 12
Reposted by Daniel Petras
Mingxun Wang
@mingxunwang.bsky.social
· May 12
Reposted by Daniel Petras
Reposted by Daniel Petras
Daniel Petras
@daniel-petras.bsky.social
· Apr 13
Reposted by Daniel Petras
Daniel Petras
@daniel-petras.bsky.social
· Mar 31
Reposted by Daniel Petras
Mingxun Wang
@mingxunwang.bsky.social
· Mar 5
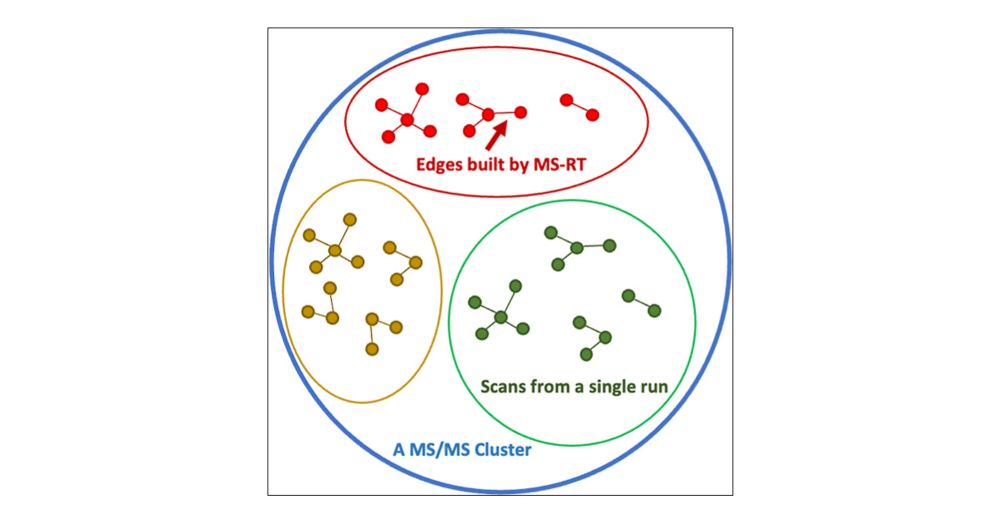
MS-RT: A Method for Evaluating MS/MS Clustering Performance for Metabolomics Data
The clustering of tandem mass spectra (MS/MS) is a crucial computational step to deduplicate repeated acquisitions in data-dependent experiments. This technique is essential in untargeted metabolomics, particularly with high-throughput mass spectrometers capable of generating hundreds of MS/MS spectra per second. Despite advancements in MS/MS clustering algorithms in proteomics, their performance in metabolomics has not been extensively evaluated due to the lack of database search tools with false discovery rate control for molecule identification. To bridge this gap, this study introduces the MS1-retention time (MS-RT) method to assess MS/MS clustering performance in metabolomics data sets. Here, we validate MS-RT by comparing MS-RT to established proteomics clustering evaluation approaches that utilize database search identifications. Additionally, we evaluate the performance of several MS/MS clustering tools on metabolomics data sets, highlighting their advantages and drawbacks. This MS-RT method and the MS/MS clustering tool benchmarking will provide valuable real world practical recommendations for tools and set the stage for future advancements in metabolomics MS/MS clustering.
doi.org