Erich M. Schwarz
@erichmschwarz.bsky.social
140 followers
91 following
15 posts
Molecular biologist using functional genomics. Started with C. elegans, then diversified.
Posts
Media
Videos
Starter Packs
Reposted by Erich M. Schwarz
Gavin Woodruff
@weirdworms.bsky.social
· Aug 23
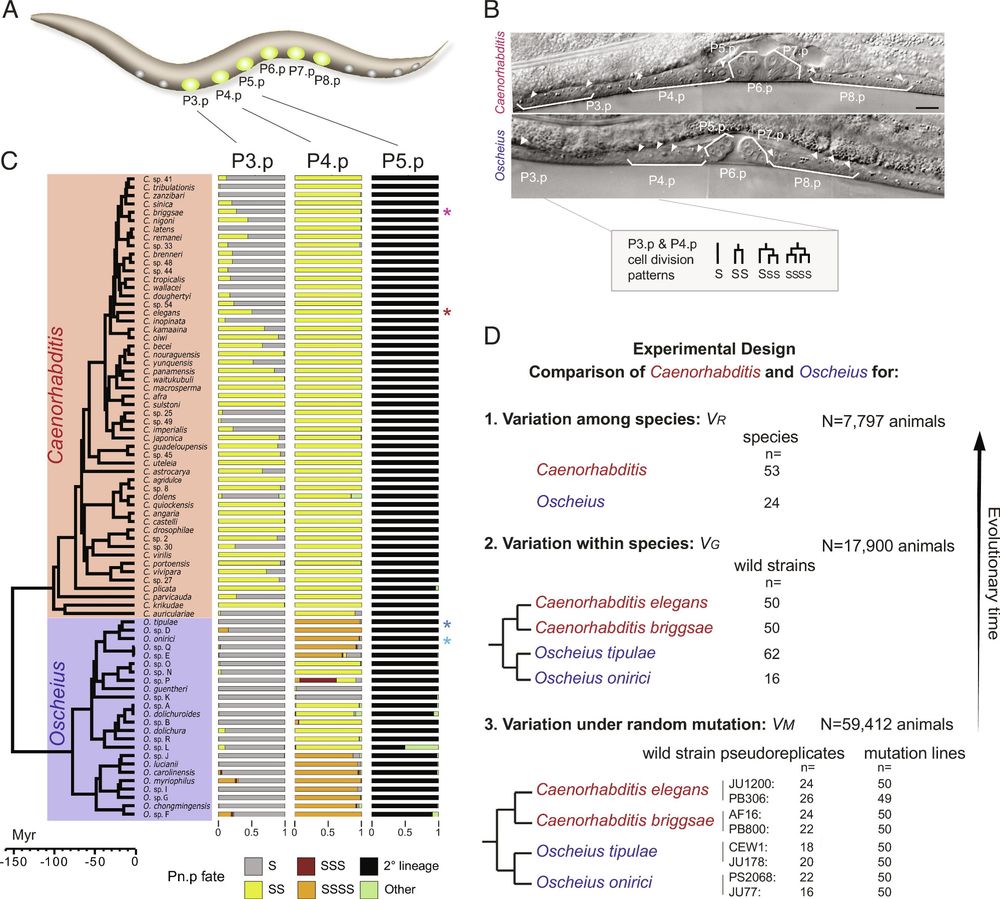
Evolution of developmental bias explains divergent patterns of phenotypic evolution in two nematode clades | PNAS
Rates of phenotypic evolution vary across traits, and these evolutionary patterns
themselves evolve. Understanding how development contributes to s...
www.pnas.org
Reposted by Erich M. Schwarz