Ernst Schmid
@ernstschmid.bsky.social
100 followers
160 following
6 posts
Full stack developer who occasionally wears a lab coat. Graduate student at Harvard Medical School.
Posts
Media
Videos
Starter Packs
Reposted by Ernst Schmid
Ernst Schmid
@ernstschmid.bsky.social
· Aug 16
Reposted by Ernst Schmid
Reposted by Ernst Schmid
Reposted by Ernst Schmid
Reposted by Ernst Schmid
anna brogan
@apbrogan.bsky.social
· Jun 17
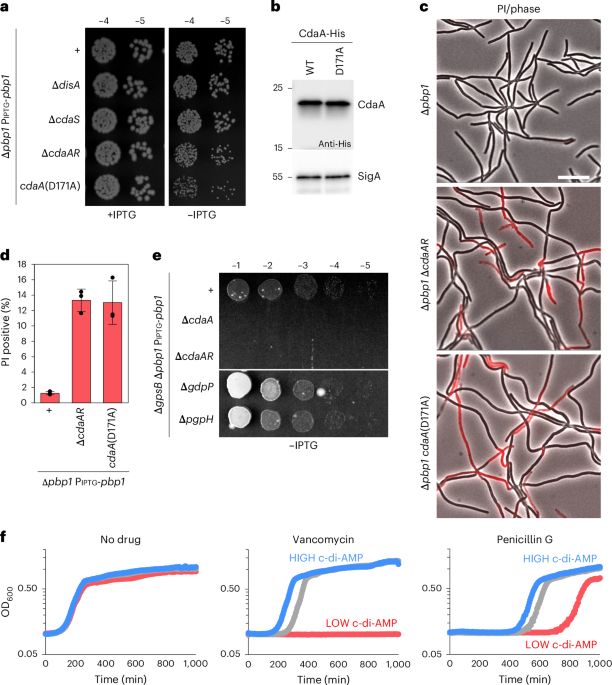
Cyclic-di-AMP modulates cellular turgor in response to defects in bacterial cell wall synthesis - Nature Microbiology
Brogan et al. uncover a signalling pathway in which levels of the nucleotide second messenger c-di-AMP increase in response to defects in cell wall synthesis. This regulatory pathway decreases turgor ...
www.nature.com
Ernst Schmid
@ernstschmid.bsky.social
· May 28

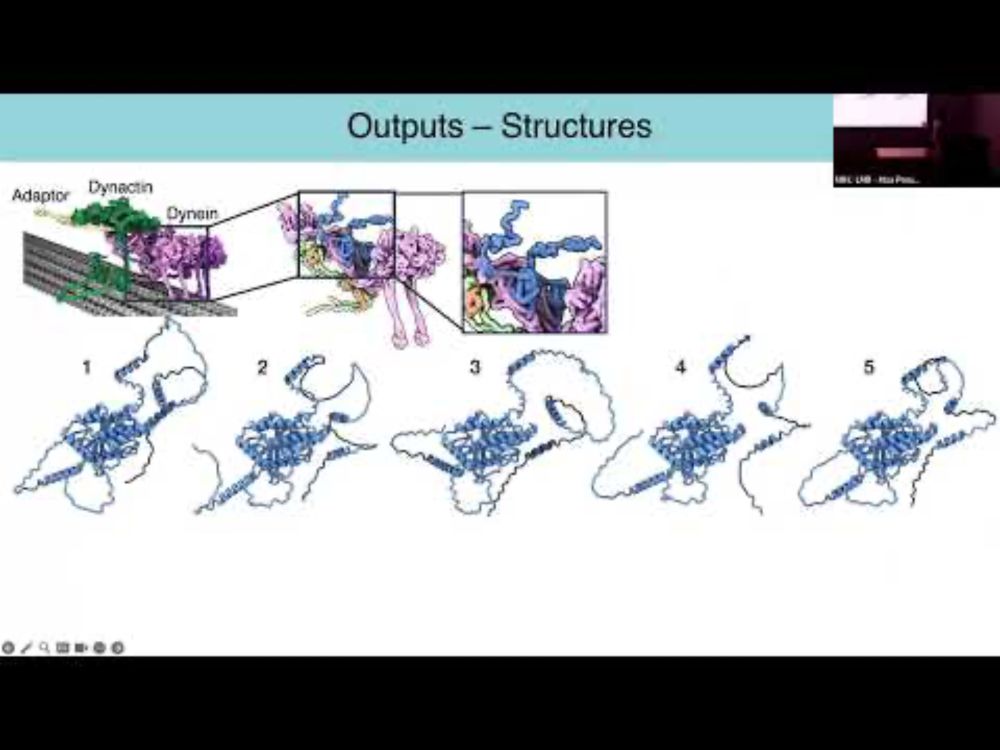
EndoMAP.v1 charts the structural landscape of human early endosome complexes - Nature
A study presents EndoMAP.v1, a resource that combines information on protein interactions and crosslink-supported structural predictions to map the interaction landscape of early endosomes.
www.nature.com
Reposted by Ernst Schmid
Joe Loparo
@joeloparo.bsky.social
· May 21
Reposted by Ernst Schmid
Reposted by Ernst Schmid
Nick Polizzi
@nickpolizzi.bsky.social
· Apr 28

Zero-shot design of drug-binding proteins via neural selection-expansion
Computational design of molecular recognition remains challenging despite advances in deep learning. The design of proteins that bind to small molecules has been particularly difficult because it requ...
www.biorxiv.org
Reposted by Ernst Schmid
Reposted by Ernst Schmid
Reposted by Ernst Schmid
Reposted by Ernst Schmid
Reposted by Ernst Schmid
Reposted by Ernst Schmid
Reposted by Ernst Schmid
Reposted by Ernst Schmid
Ernst Schmid
@ernstschmid.bsky.social
· Feb 26
Ernst Schmid
@ernstschmid.bsky.social
· Feb 26
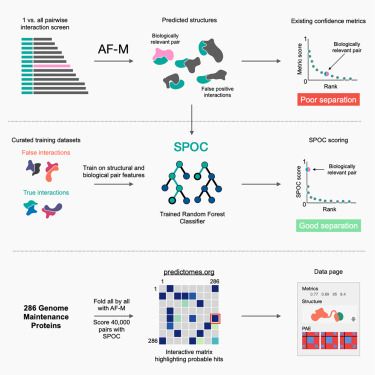
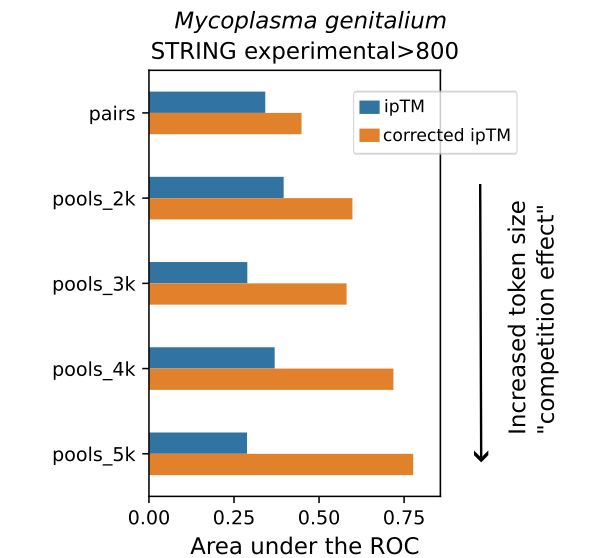
Predictomes, a classifier-curated database of AlphaFold-modeled protein-protein interactions
Schmid and Walter train a classifier that discerns functionally relevant structure
predictions in proteome-wide protein-protein interaction (PPI) screens using AlphaFold-Multimer,
and they use this co...
www.cell.com