Laboratory of Functional Viromics (LETKO)
@fviromics.bsky.social
1.3K followers
280 following
63 posts
Dr. Michael Letko’s Laboratory of Functional Viromics | we study zoonotic potential of novel viruses.
For more information, check out our website: https://labs.wsu.edu/lofv/
Posts
Media
Videos
Starter Packs
Reposted by Laboratory of Functional Viromics (LETKO)
Reposted by Laboratory of Functional Viromics (LETKO)
Reposted by Laboratory of Functional Viromics (LETKO)
Reposted by Laboratory of Functional Viromics (LETKO)
Reposted by Laboratory of Functional Viromics (LETKO)
Reposted by Laboratory of Functional Viromics (LETKO)
Reposted by Laboratory of Functional Viromics (LETKO)
Reposted by Laboratory of Functional Viromics (LETKO)
Reposted by Laboratory of Functional Viromics (LETKO)
Reposted by Laboratory of Functional Viromics (LETKO)
Reposted by Laboratory of Functional Viromics (LETKO)
Reposted by Laboratory of Functional Viromics (LETKO)
Reposted by Laboratory of Functional Viromics (LETKO)
Reposted by Laboratory of Functional Viromics (LETKO)
Reposted by Laboratory of Functional Viromics (LETKO)
Arinjay Banerjee
@banerjeelab.ca
· Jul 1
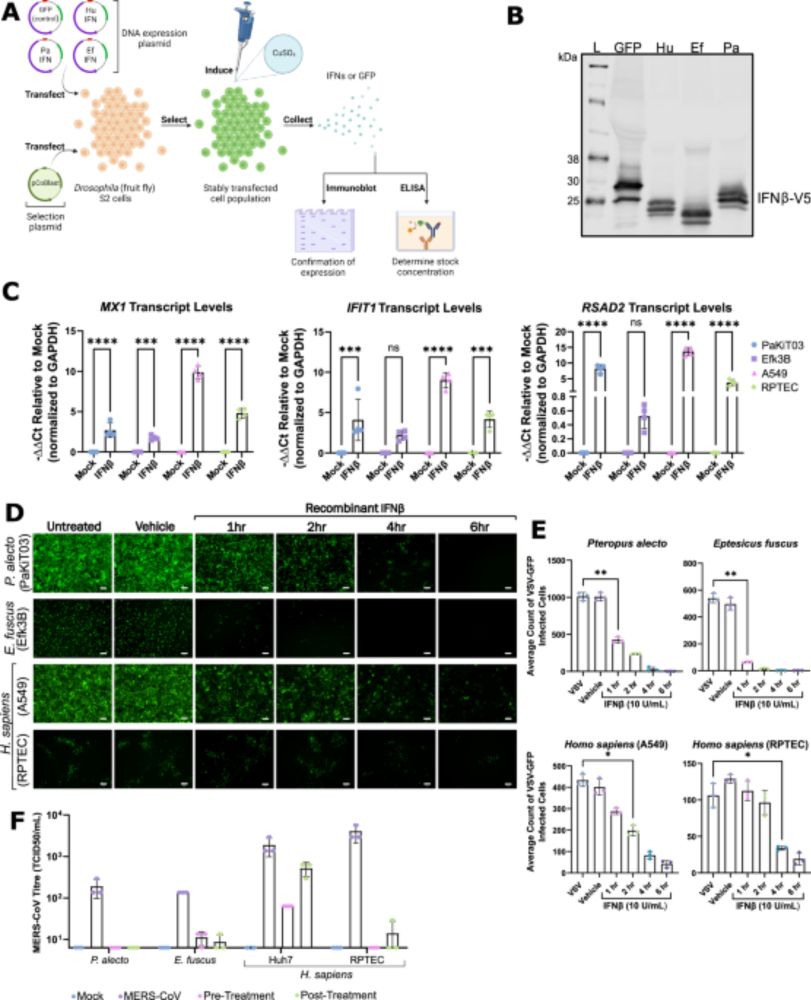
Bat-specific adaptations in interferon signaling and GBP1 contribute to enhanced antiviral capacity - Nature Communications
Bats harbor diverse viruses but it’s less clear how they tolerate infection. Here, by characterizing innate immune responses in bat cells the authors show that IFN-beta signaling resists antagonistic ...
www.nature.com