André Soares
@geomicrosoares.bsky.social
500 followers
790 following
31 posts
🇵🇹 Staff Scientist @probstlab.bsky.social
(@unidue.bsky.social - 🇩🇪). Microbial genomics in the One Health context, biogeochemistry of cave microbiomes, alga-microbe symbioses.
Posts
Media
Videos
Starter Packs
Reposted by André Soares
Reposted by André Soares
Reposted by André Soares
Reposted by André Soares
Reposted by André Soares
Reposted by André Soares
Reposted by André Soares
Reposted by André Soares
Reposted by André Soares
Vlad Bondarenko
@vladbndk.bsky.social
· Sep 5
Reposted by André Soares
Reposted by André Soares
Arwyn Edwards
@arwynedwards.bsky.social
· Aug 27
Reposted by André Soares
Maria Dzunkova
@dzunkovam.bsky.social
· Aug 26
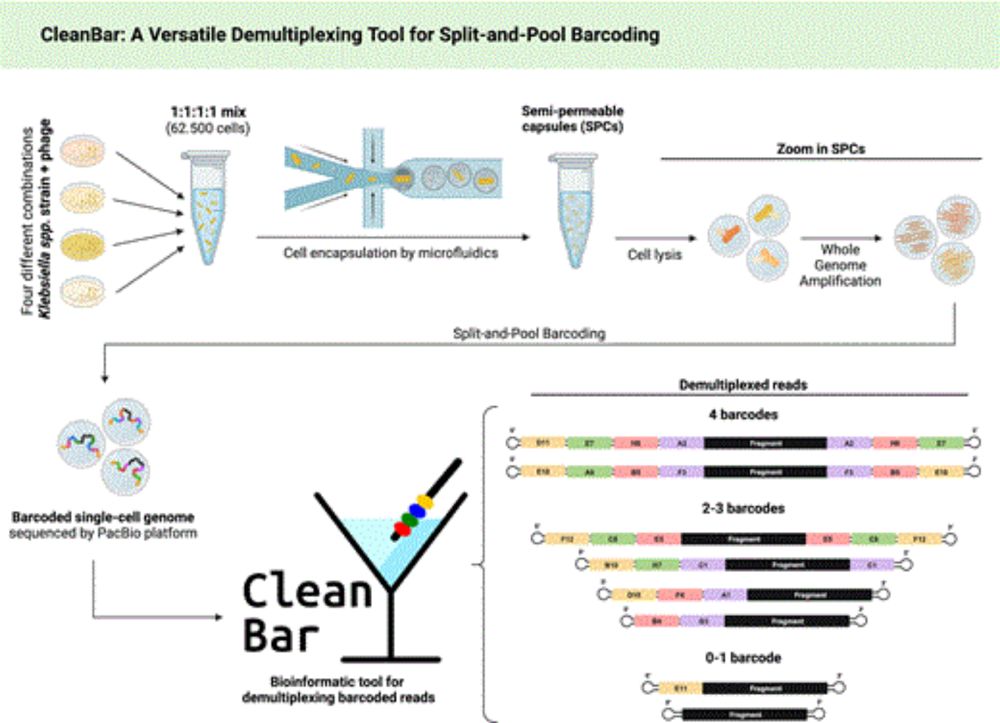
CleanBar: a versatile demultiplexing tool for split-and-pool barcoding in single-cell omics
Abstract. Split-and-pool barcoding generates thousands of unique barcode strings through sequential ligations in 96-well plates, making single-cell omics m
academic.oup.com
Reposted by André Soares
Cameron Thrash
@jcamthrash.bsky.social
· Aug 20

A novel bacterial protein family that catalyses nitrous oxide reduction - Nature
Cultivation of tropical soil microorganisms combined with physiological experiments and bioinformatics analyses identify a family of clade III lactonase-type nitrous oxide reductases with low sequence...
www.nature.com
Reposted by André Soares
Taylor priest
@taylorpriest.bsky.social
· Aug 14

The JEDI marker as a universal measure of planetary biodiversity
Despite its critical importance in the formation and maintenance of ecosystems and homeostasis on Earth, biodiversity remains a complex and non-unified concept. Consequently, standards for measuring b...
www.biorxiv.org