Jana Helsen
@helsenjana.bsky.social
1.5K followers
870 following
39 posts
Evolutionary cell biology, chromosomes, yeasts, and occasional SciArt🧬🧪🎨. Postdoctoral fellow at the labs of Gautam Dey (EMBL) and Gavin Sherlock (Stanford University).
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Jana Helsen
Gerlich Lab
@gerlichlab.bsky.social
· Sep 3

An electrostatic repulsion model of centromere organisation
During cell division, chromosomes reorganise into compact bodies in which centromeres localise precisely at the chromatin surface to enable kinetochore-microtubule interactions essential for genome se...
doi.org
Reposted by Jana Helsen
Reposted by Jana Helsen
Reposted by Jana Helsen
Tera Levin
@teralevin.bsky.social
· Aug 5

Hypermutable hotspot enables the rapid evolution of self/non-self recognition genes in Dictyostelium
Cells require highly polymorphic receptors to perform accurate self/non-self recognition. In the amoeba Dicytostelium discoideum, polymorphic TgrB1 & TgrC1 proteins are used to bind sister cells and e...
www.biorxiv.org
Reposted by Jana Helsen
Marko Kaksonen
@kaksonen.bsky.social
· Jul 10
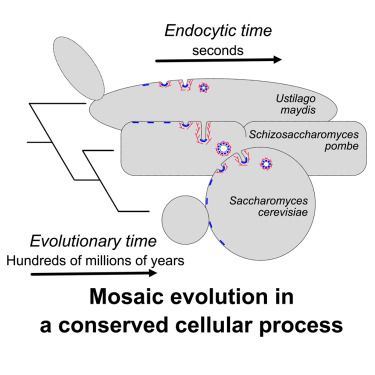
Mosaic evolution of clathrin-mediated endocytosis in fungi
Picco et al. take an evolutionary approach to study clathrin-mediated endocytosis
in a fungal clade and discover that the process is evolving in a mosaic fashion. The
early phase of endocytosis has sh...
www.cell.com
Reposted by Jana Helsen
Max Haase
@maxhaase.bsky.social
· Apr 25

Ancient co-option of LTR retrotransposons as yeast centromeres
The evolutionary origins of the genetic point centromere in the brewer's yeast Saccharomyces cerevisiae, a member of the order Saccharomycetales, are still unknown. Competing hypotheses suggest that t...
www.biorxiv.org
Reposted by Jana Helsen
Jana Helsen
@helsenjana.bsky.social
· Apr 3
Reposted by Jana Helsen
Hashim
@hashimreza.bsky.social
· Apr 3

Autophagy-related protein Atg11 is essential for microtubule-mediated chromosome segregation
Autophagy-related proteins have roles in several cellular processes, but their function in genome stability remains unclear. Here, the authors show that Atg11 has an essential non-canonical function i...
doi.org
Jana Helsen
@helsenjana.bsky.social
· Jan 17
Jana Helsen
@helsenjana.bsky.social
· Jan 17
Jana Helsen
@helsenjana.bsky.social
· Jan 17
Jana Helsen
@helsenjana.bsky.social
· Jan 17
Jana Helsen
@helsenjana.bsky.social
· Jan 17
Jana Helsen
@helsenjana.bsky.social
· Jan 17
Jana Helsen
@helsenjana.bsky.social
· Jan 17
Jana Helsen
@helsenjana.bsky.social
· Dec 26
Reposted by Jana Helsen