Maric Lab
@hmariclab.bsky.social
180 followers
280 following
7 posts
#EmmyNoether Lab of Hans Maric @uni-wuerzburg.de
#ChemBio #Peptides #PNA #Microarrays #ChemicalProbes pharmacological targeting of #PPI #IDR #RNA
http://MaricLab.com
https://www.uni-wuerzburg.de/en/rvz/research-groups/maric-group/
http://bit.ly/14S4Z8k.
Posts
Media
Videos
Starter Packs
Pinned
Maric Lab
@hmariclab.bsky.social
· May 5
Maric Lab
@hmariclab.bsky.social
· May 14
Reposted by Maric Lab
Reposted by Maric Lab
Reposted by Maric Lab
Reposted by Maric Lab
Malte Gersch
@maltegersch.bsky.social
· May 5
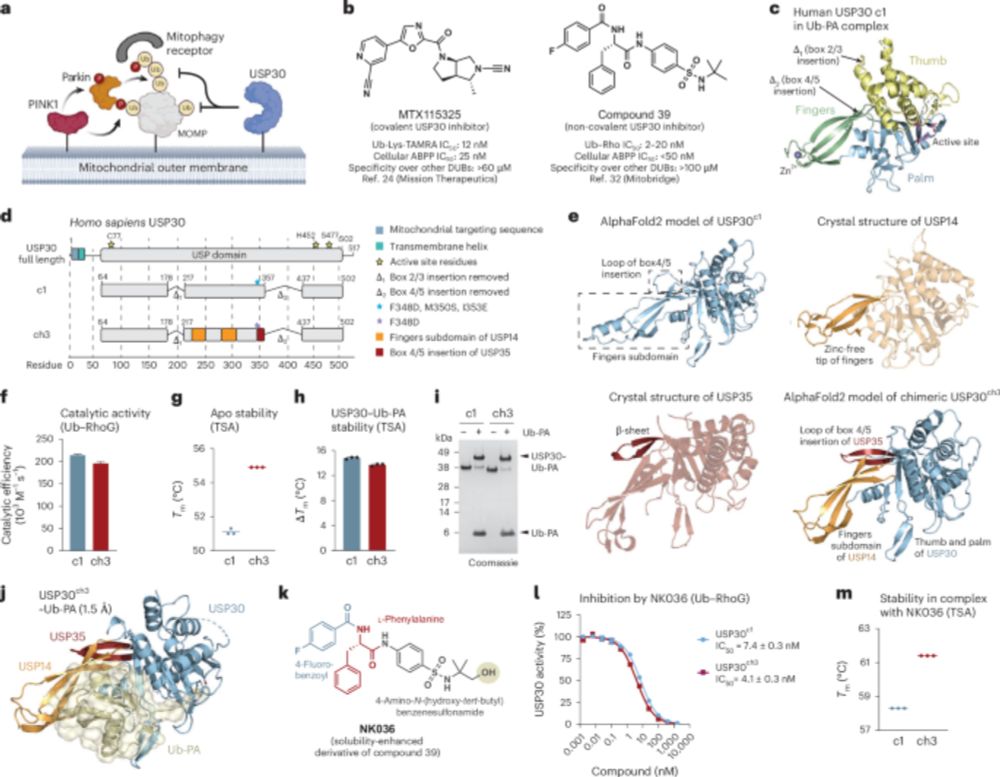
Chimeric deubiquitinase engineering reveals structural basis for specific inhibition of the mitophagy regulator USP30 - Nature Structural & Molecular Biology
Kazi et al. report the crystal structure of the mitochondrial deubiquitinase USP, a clinical stage Parkinson’s disease drug target, in complex with a specific inhibitor. The authors delineate a framew...
www.nature.com
Reposted by Maric Lab
Maric Lab
@hmariclab.bsky.social
· May 5
Reposted by Maric Lab
Reposted by Maric Lab
Sauer Lab
@sauer-lab.bsky.social
· Feb 12
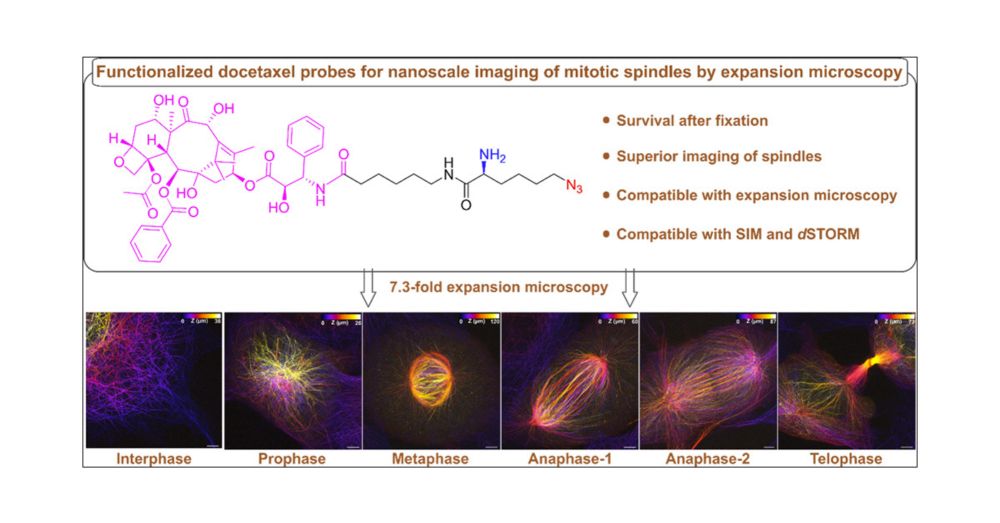
Functionalized Docetaxel Probes for Refined Visualization of Mitotic Spindles by Expansion Microscopy
Visualizing the ultrastructure of mitotic spindles, the macromolecular machines that segregate chromosomes during mitosis, by fluorescence imaging remains challenging. Here we introduce an azido- and ...
pubs.acs.org
Reposted by Maric Lab
Maric Lab
@hmariclab.bsky.social
· Mar 20
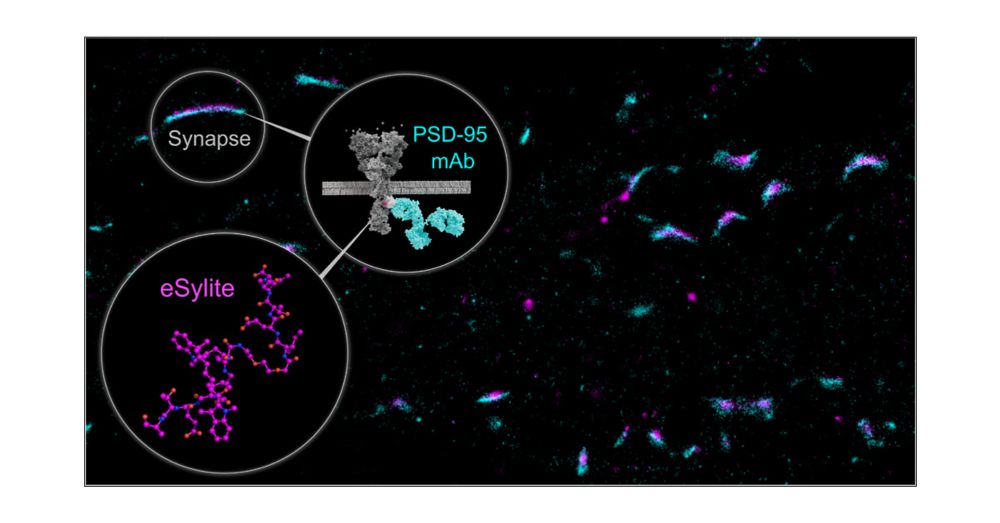
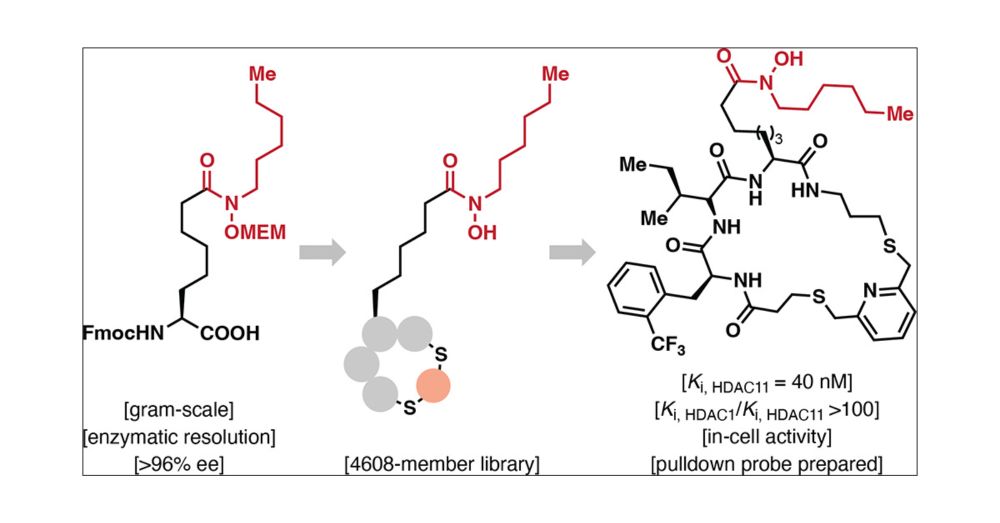
eSylites: Synthetic Probes for Visualization and Topographic Mapping of Single Excitatory Synapses
The spatiotemporal organization of the postsynaptic density (PSD) is a fundamental determinant of synaptic transmission, information processing, and storage in the brain. The major bottleneck that prevents the direct and precise representation of the nanometer-scaled organization of excitatory glutamatergic synapses is the size of antibodies, nanobodies, and the genetically encoded fluorescent tags. Here, we introduce small, high affinity synthetic probes for simplified, high contrast visualization of excitatory synapses without the limitations of larger biomolecules. In vitro binding quantification together with microscopy-based evaluation identified eSylites, a series of fluorescent bivalent peptides comprising a dye, linker, and sequence composition that show remarkable cellular target selectivity. Applied on primary neurons or brain slices at nanomolar concentrations, eSylites specifically report PSD-95, the key orchestrator of glutamate receptor nanodomains juxtaposed to the presynaptic glutamate release sites that mediate fast synaptic transmission. The eSylite design minimizes a spatial dye offset and thereby enables visualization of PSD-95 with improved localization precision and further time-resolved discrimination. In particular, we find that individual dendritic spines can contain separate nanodomains enriched for either PSD-95 or its closest homologues, PSD-93 or SAP102. Collectively, these data establish eSylites as a broadly applicable tool for simplified excitatory synapse visualization, as well as a high-end microscopy compatible probe for resolving the PSD organization with unprecedented resolution.
pubs.acs.org
Reposted by Maric Lab
Maric Lab
@hmariclab.bsky.social
· Mar 20

eSylites: Synthetic Probes for Visualization and Topographic Mapping of Single Excitatory Synapses
The spatiotemporal organization of the postsynaptic density (PSD) is a fundamental determinant of synaptic transmission, information processing, and storage in the brain. The major bottleneck that prevents the direct and precise representation of the nanometer-scaled organization of excitatory glutamatergic synapses is the size of antibodies, nanobodies, and the genetically encoded fluorescent tags. Here, we introduce small, high affinity synthetic probes for simplified, high contrast visualization of excitatory synapses without the limitations of larger biomolecules. In vitro binding quantification together with microscopy-based evaluation identified eSylites, a series of fluorescent bivalent peptides comprising a dye, linker, and sequence composition that show remarkable cellular target selectivity. Applied on primary neurons or brain slices at nanomolar concentrations, eSylites specifically report PSD-95, the key orchestrator of glutamate receptor nanodomains juxtaposed to the presynaptic glutamate release sites that mediate fast synaptic transmission. The eSylite design minimizes a spatial dye offset and thereby enables visualization of PSD-95 with improved localization precision and further time-resolved discrimination. In particular, we find that individual dendritic spines can contain separate nanodomains enriched for either PSD-95 or its closest homologues, PSD-93 or SAP102. Collectively, these data establish eSylites as a broadly applicable tool for simplified excitatory synapse visualization, as well as a high-end microscopy compatible probe for resolving the PSD organization with unprecedented resolution.
pubs.acs.org
Maric Lab
@hmariclab.bsky.social
· Mar 20

eSylites: Synthetic Probes for Visualization and Topographic Mapping of Single Excitatory Synapses
The spatiotemporal organization of the postsynaptic density (PSD) is a fundamental determinant of synaptic transmission, information processing, and storage in the brain. The major bottleneck that prevents the direct and precise representation of the nanometer-scaled organization of excitatory glutamatergic synapses is the size of antibodies, nanobodies, and the genetically encoded fluorescent tags. Here, we introduce small, high affinity synthetic probes for simplified, high contrast visualization of excitatory synapses without the limitations of larger biomolecules. In vitro binding quantification together with microscopy-based evaluation identified eSylites, a series of fluorescent bivalent peptides comprising a dye, linker, and sequence composition that show remarkable cellular target selectivity. Applied on primary neurons or brain slices at nanomolar concentrations, eSylites specifically report PSD-95, the key orchestrator of glutamate receptor nanodomains juxtaposed to the presynaptic glutamate release sites that mediate fast synaptic transmission. The eSylite design minimizes a spatial dye offset and thereby enables visualization of PSD-95 with improved localization precision and further time-resolved discrimination. In particular, we find that individual dendritic spines can contain separate nanodomains enriched for either PSD-95 or its closest homologues, PSD-93 or SAP102. Collectively, these data establish eSylites as a broadly applicable tool for simplified excitatory synapse visualization, as well as a high-end microscopy compatible probe for resolving the PSD organization with unprecedented resolution.
pubs.acs.org
Reposted by Maric Lab
Reposted by Maric Lab
Ivan Talucci
@ivantalucci.bsky.social
· Mar 4
Maric Lab
@hmariclab.bsky.social
· Jan 28
Reposted by Maric Lab
Reposted by Maric Lab
Reposted by Maric Lab