Danny IncaRNAto
@incarnatolab.bsky.social
930 followers
650 following
43 posts
Associate Professor in Molecular Genetics | University of Groningen | Chief R&D Officer @ Serna Bio | #RNA structural ensembles & dynamics | https://www.incaRNAtolab.com | Views are my own
Posts
Media
Videos
Starter Packs
Danny IncaRNAto
@incarnatolab.bsky.social
· Jul 30
Danny IncaRNAto
@incarnatolab.bsky.social
· Jul 29
Danny IncaRNAto
@incarnatolab.bsky.social
· Jul 29
Danny IncaRNAto
@incarnatolab.bsky.social
· Jul 28
Danny IncaRNAto
@incarnatolab.bsky.social
· Jul 27
Danny IncaRNAto
@incarnatolab.bsky.social
· Jul 27
Danny IncaRNAto
@incarnatolab.bsky.social
· Jul 27
Danny IncaRNAto
@incarnatolab.bsky.social
· Jul 27
Danny IncaRNAto
@incarnatolab.bsky.social
· Jul 27
Danny IncaRNAto
@incarnatolab.bsky.social
· Jul 27
Danny IncaRNAto
@incarnatolab.bsky.social
· Jul 27
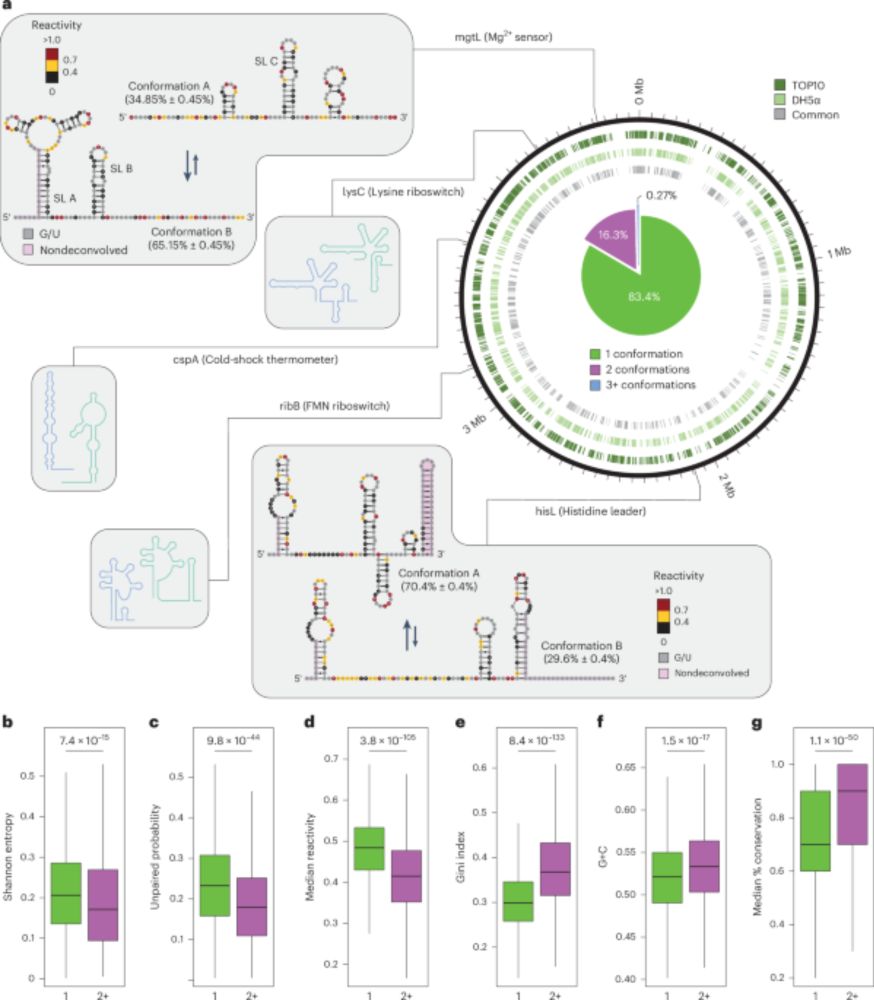
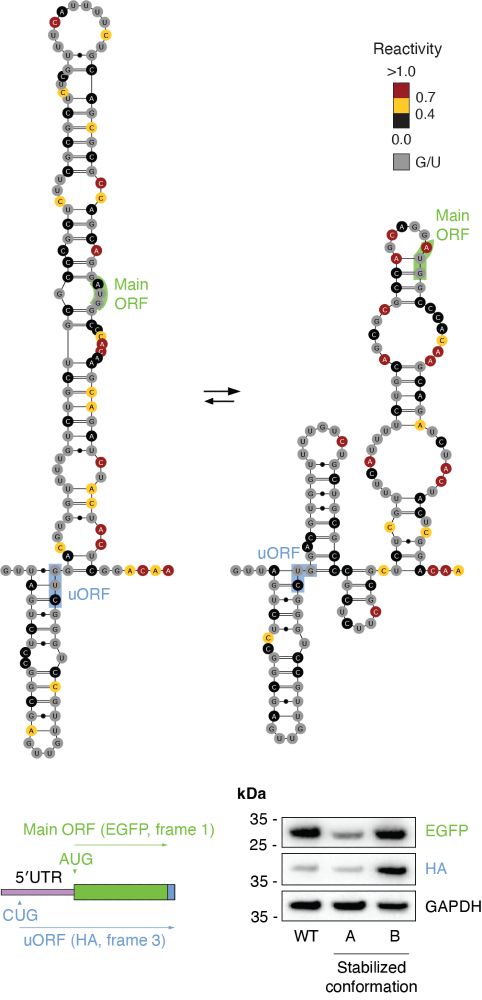
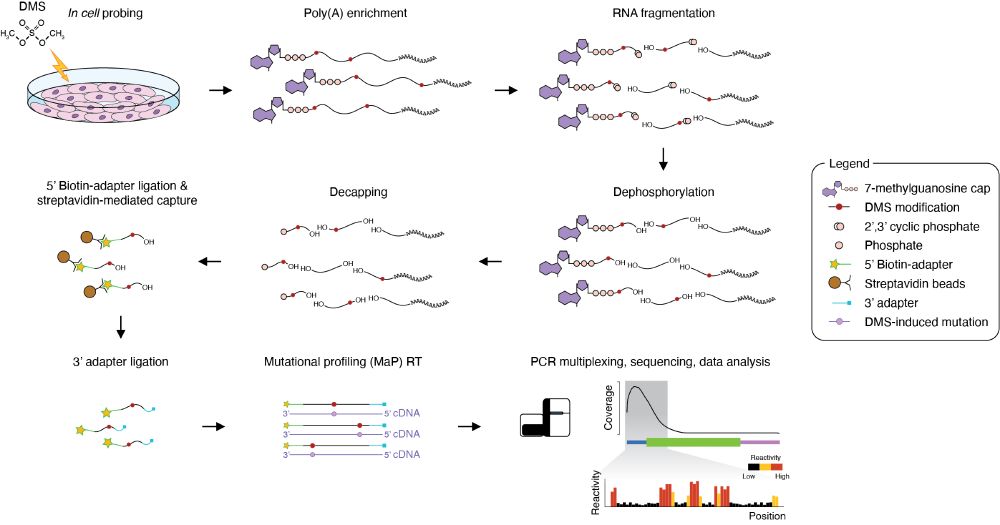
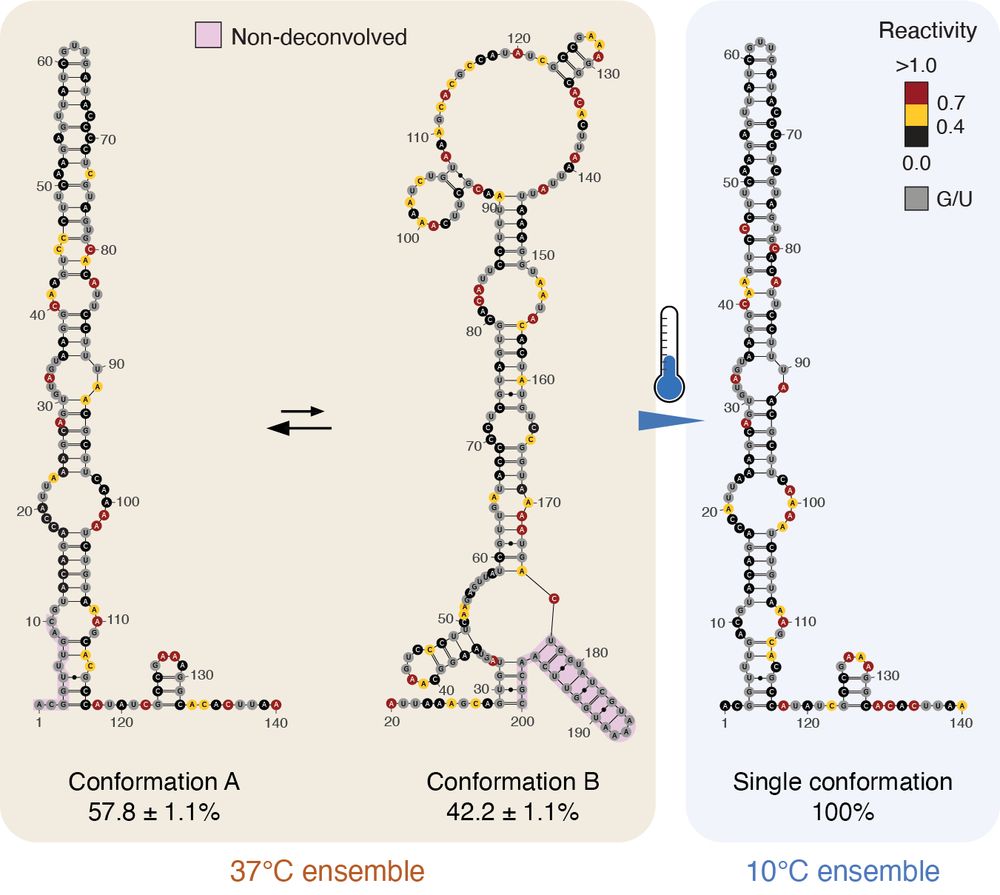
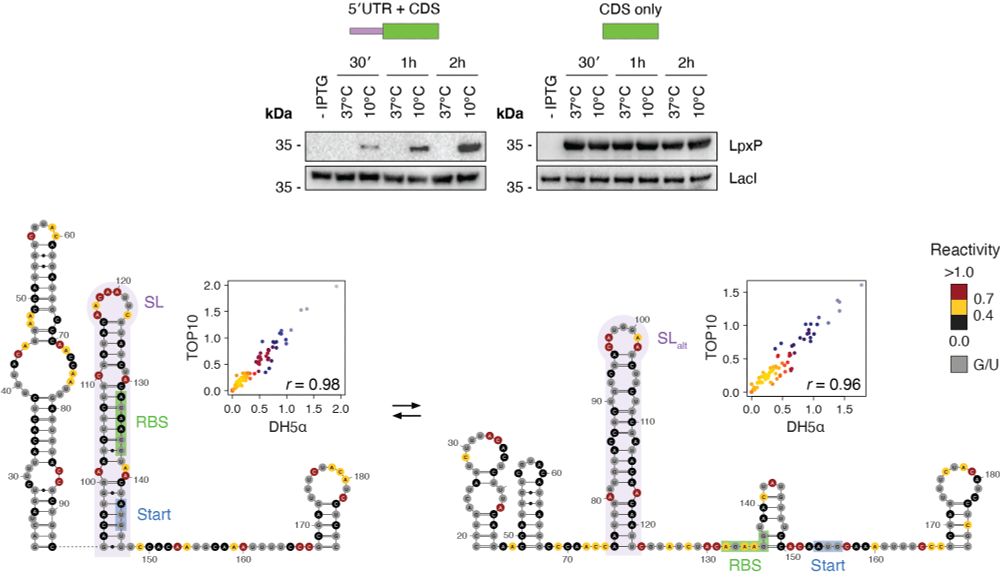
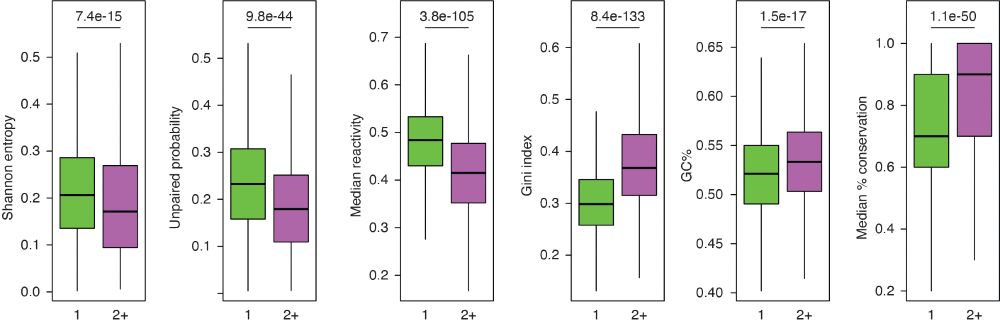
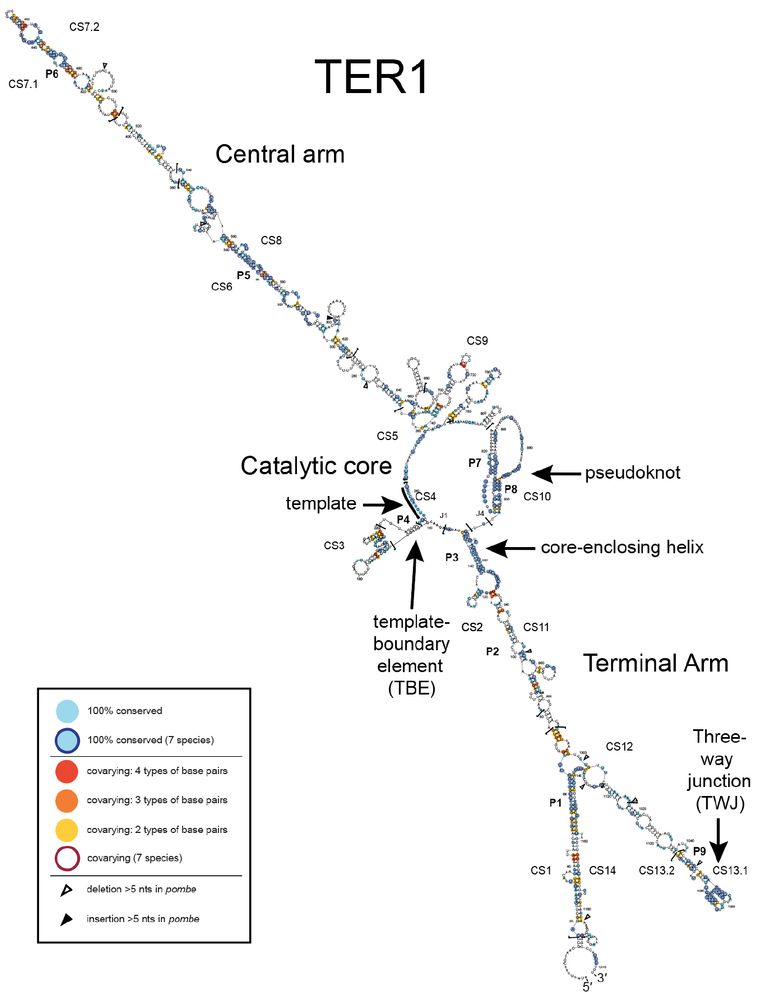
Identification of conserved RNA regulatory switches in living cells using RNA secondary structure ensemble mapping and covariation analysis - Nature Biotechnology
Transcriptome-scale maps of RNA secondary structure ensembles in living cells detect candidate RNA structural switches.
nature.com
Danny IncaRNAto
@incarnatolab.bsky.social
· Jun 19
Reposted by Danny IncaRNAto
Reposted by Danny IncaRNAto
Sean Eddy
@cryptogenomicon.bsky.social
· May 22
Reposted by Danny IncaRNAto