Inês Cunha
@inesmcunha.bsky.social
170 followers
110 following
18 posts
Biomedical Engineer 🤓 Now PhD student at the CMCB Lab, SciLifeLab. Learning how to gain knowledge from images 🦠🔬with AI 💻
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Inês Cunha
Reposted by Inês Cunha
Sebastian Bauer
@sebjbauer.bsky.social
· Aug 29
Reposted by Inês Cunha
Reposted by Inês Cunha
Reposted by Inês Cunha
Inês Cunha
@inesmcunha.bsky.social
· May 14
Inês Cunha
@inesmcunha.bsky.social
· May 14

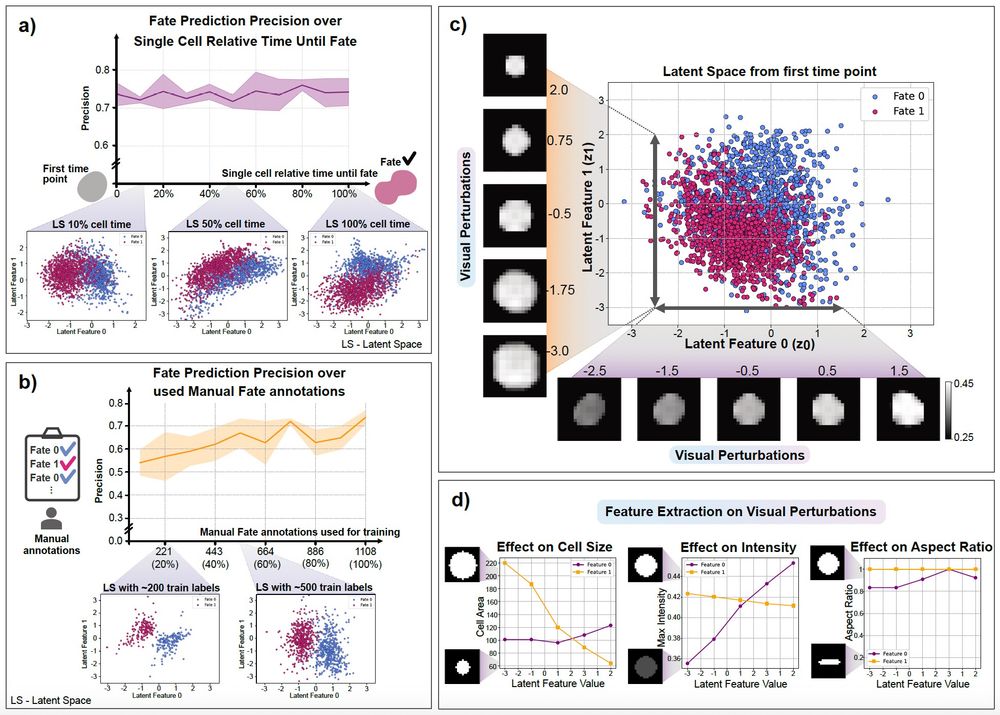
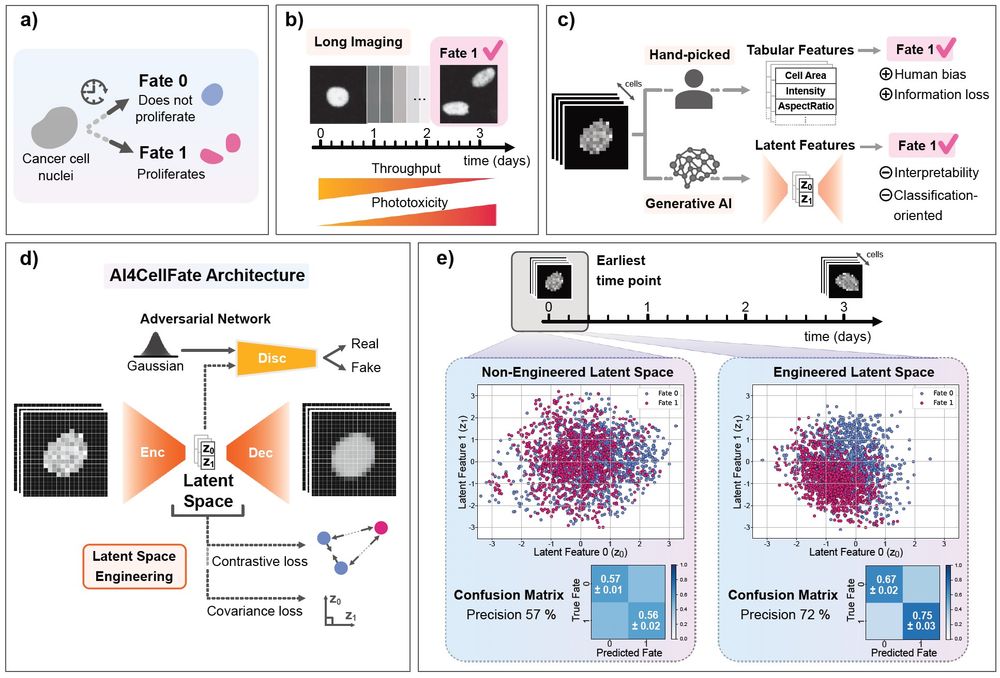
AI4CellFate: Interpretable Early Cell Fate Prediction with Generative AI
Live-cell imaging provides a unique insight into complex cellular processes including single cell fate, but remains limited by both low-throughput and the lack of generalisable analytics for the multi...
biorxiv.org
Inês Cunha
@inesmcunha.bsky.social
· May 14
Inês Cunha
@inesmcunha.bsky.social
· Feb 6