Jakob Farnung
@jakobfarnung.bsky.social
340 followers
290 following
8 posts
PostDoc Schulman Lab, MPI Biochemistry; formerly Bode Lab, ETH Zürich
Posts
Media
Videos
Starter Packs
Reposted by Jakob Farnung
Reposted by Jakob Farnung
Reposted by Jakob Farnung
Jakob Farnung
@jakobfarnung.bsky.social
· May 26
Samuel Maiwald
@samuelmaiwald.bsky.social
· May 26
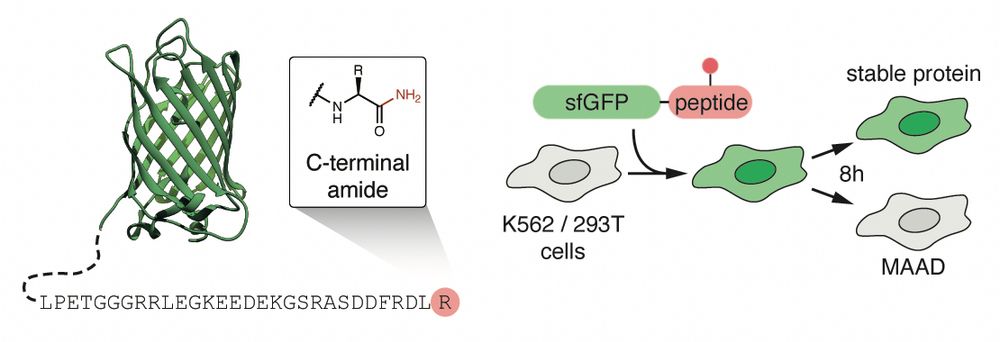
TRIP12 structures reveal HECT E3 formation of K29 linkages and branched ubiquitin chains - Nature Structural & Molecular Biology
Using biochemistry, chemical biology, and cryo-EM, Maiwald et al. elucidate how TRIP12 forms K29 linkages and K29/K48-linked branched ubiquitin chains, revealing a mechanism for polyubiquitylation sha...
www.nature.com
Reposted by Jakob Farnung
Samuel Maiwald
@samuelmaiwald.bsky.social
· May 26

TRIP12 structures reveal HECT E3 formation of K29 linkages and branched ubiquitin chains - Nature Structural & Molecular Biology
Using biochemistry, chemical biology, and cryo-EM, Maiwald et al. elucidate how TRIP12 forms K29 linkages and K29/K48-linked branched ubiquitin chains, revealing a mechanism for polyubiquitylation sha...
www.nature.com
Reposted by Jakob Farnung
Jakob Farnung
@jakobfarnung.bsky.social
· Mar 24
Leo Kiss
@leokiss.bsky.social
· Mar 24
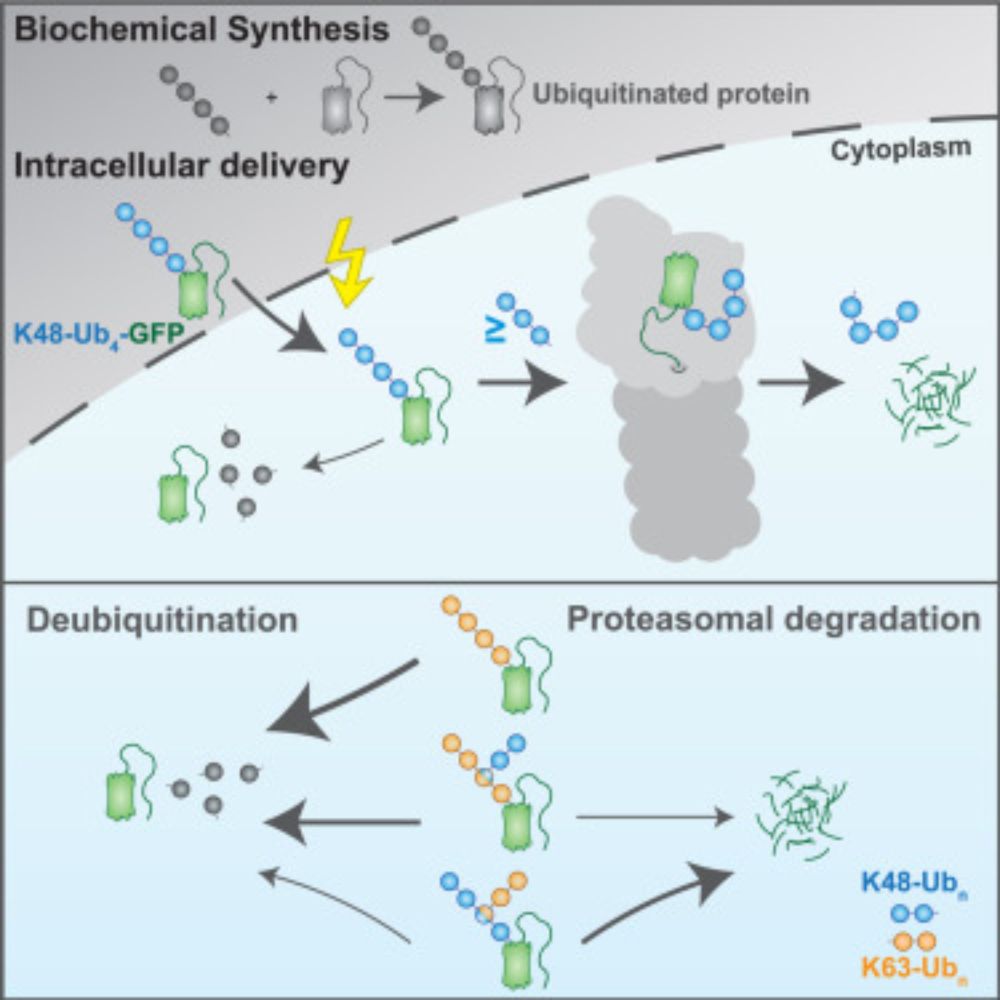
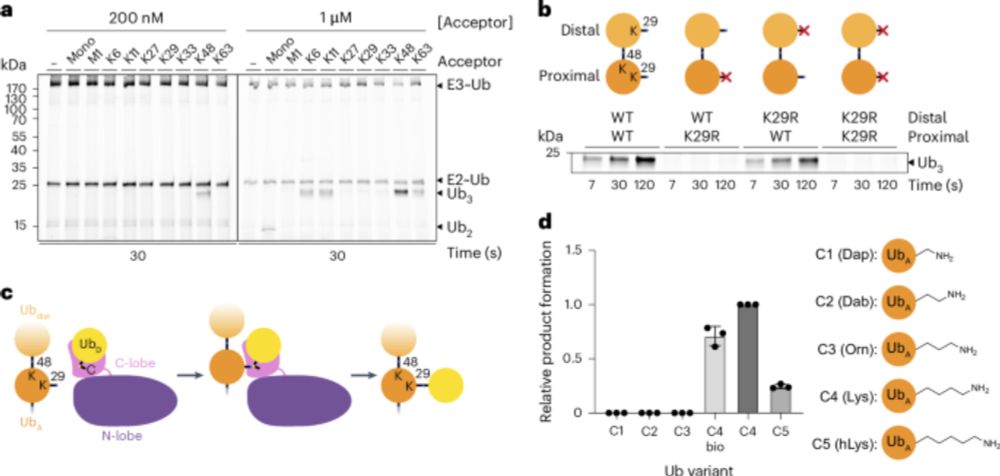
UbiREAD deciphers proteasomal degradation code of homotypic and branched K48 and K63 ubiquitin chains
Ubiquitin chains determine the fates of their modified proteins, including proteasomal
degradation. Kiss et al. present UbiREAD, a technology to monitor cellular degradation
and deubiquitination at hi...
www.cell.com
Reposted by Jakob Farnung
Reposted by Jakob Farnung
Reposted by Jakob Farnung
Lucas Farnung
@lucas.farnunglab.com
· Jan 30
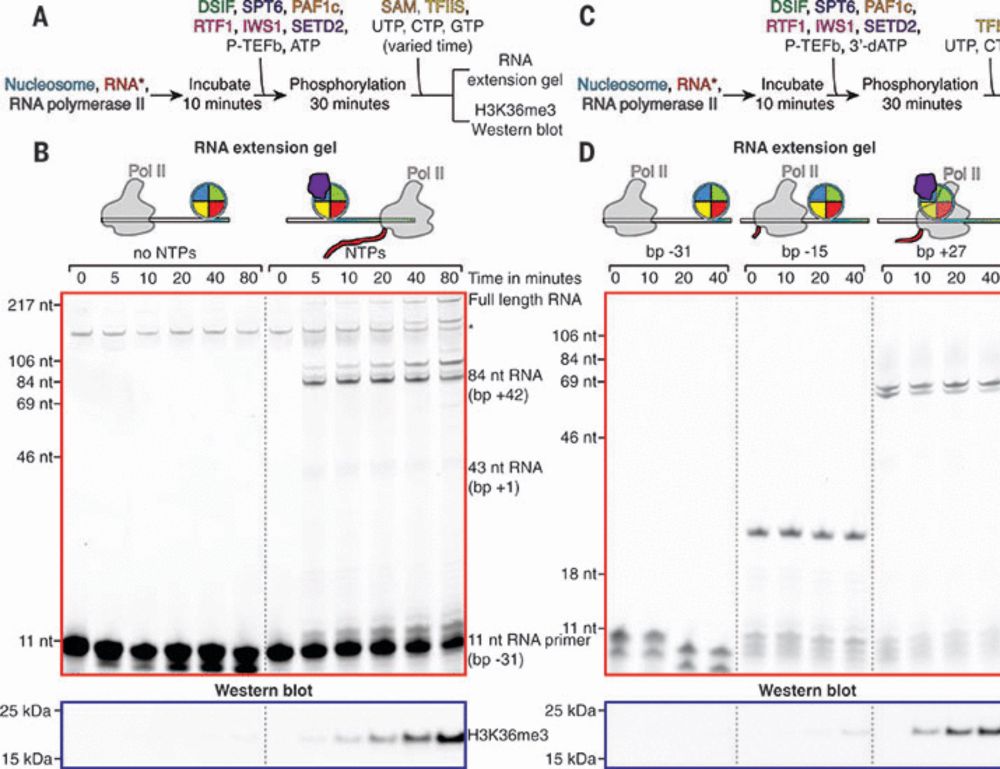
Structural basis of H3K36 trimethylation by SETD2 during chromatin transcription
During transcription, RNA polymerase II traverses through chromatin, and posttranslational modifications including histone methylations mark regions of active transcription. Histone protein H3 lysine ...
www.science.org
Reposted by Jakob Farnung
Micha Rapé Lab
@micharapelab.bsky.social
· Jan 29
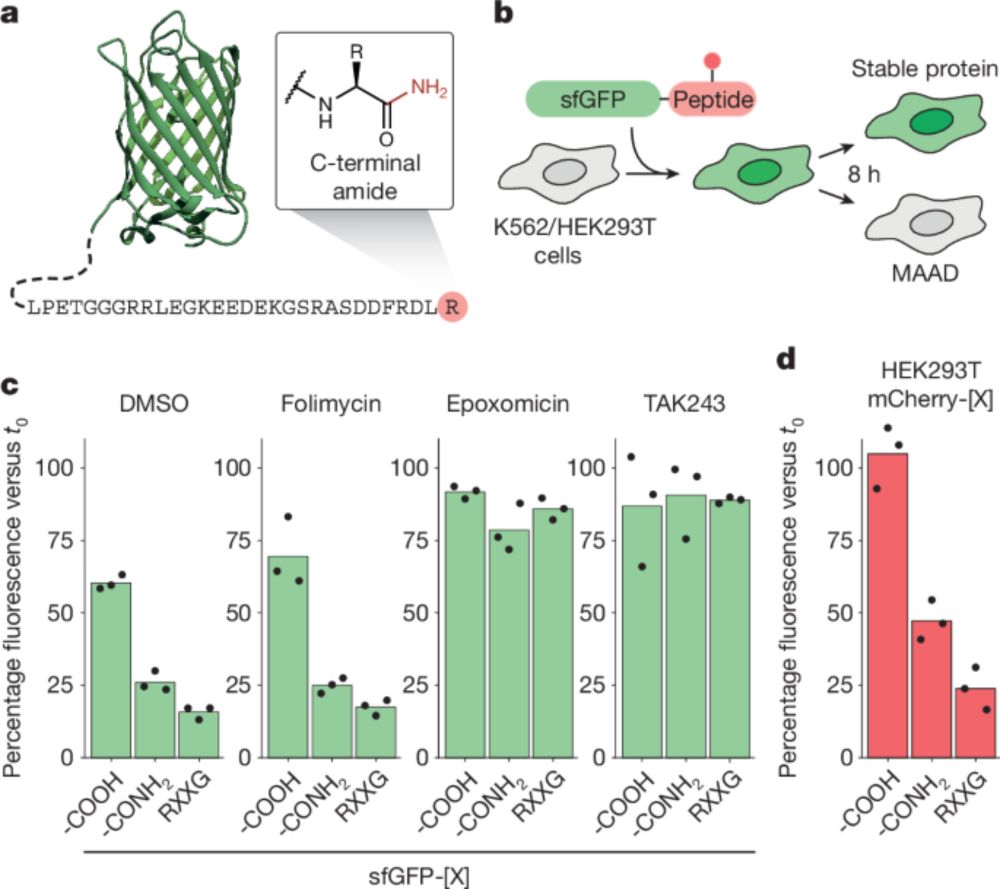
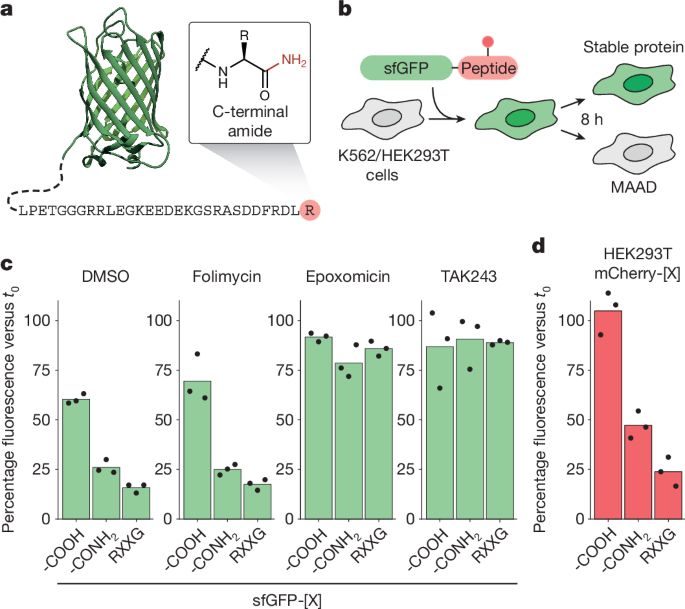
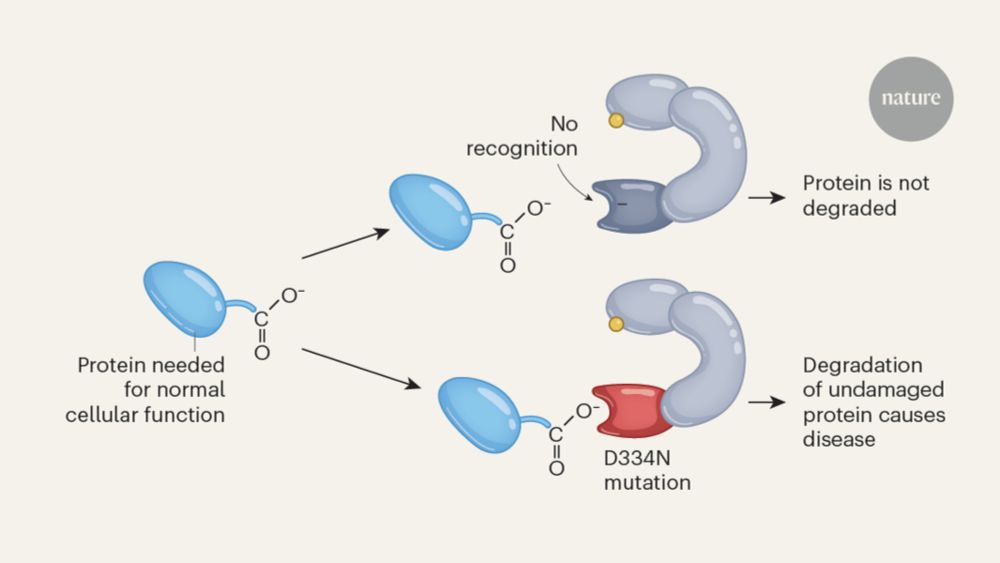
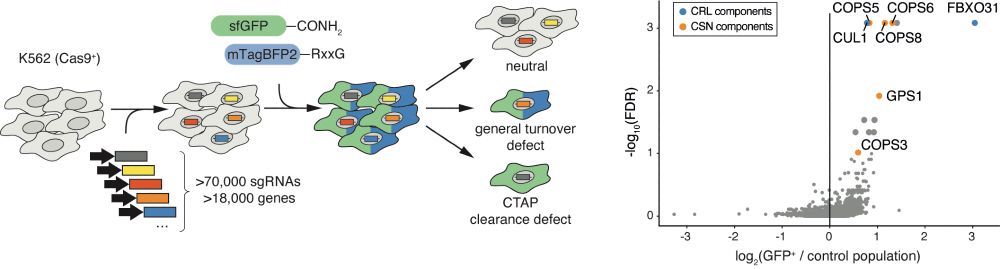
Signs of damage that drive protein degradation | Nature
Many environmental toxins damage proteins, which then must be removed to avoid dangerous protein aggregation and disease. How cells dispose of chemically modified proteins has been unclear, but a discovery offers some clues. How the stress-response machinery eliminates damaged proteins.
rdcu.be
Reposted by Jakob Farnung
Jakob Farnung
@jakobfarnung.bsky.social
· Jan 29
Jakob Farnung
@jakobfarnung.bsky.social
· Jan 29
Jakob Farnung
@jakobfarnung.bsky.social
· Jan 29