Opinions are my own and do not reflect those of my employer.
https://jbloomlab.org/
Indeed, we previously found this heterogeneity may be important for influenza evolution: elifesciences.org/reviewed-pre...

Indeed, we previously found this heterogeneity may be important for influenza evolution: elifesciences.org/reviewed-pre...
Titers are summarized below; can be examined interactively at jbloomlab.github.io/flu-seqneut-... & jbloomlab.github.io/flu-seqneut-...
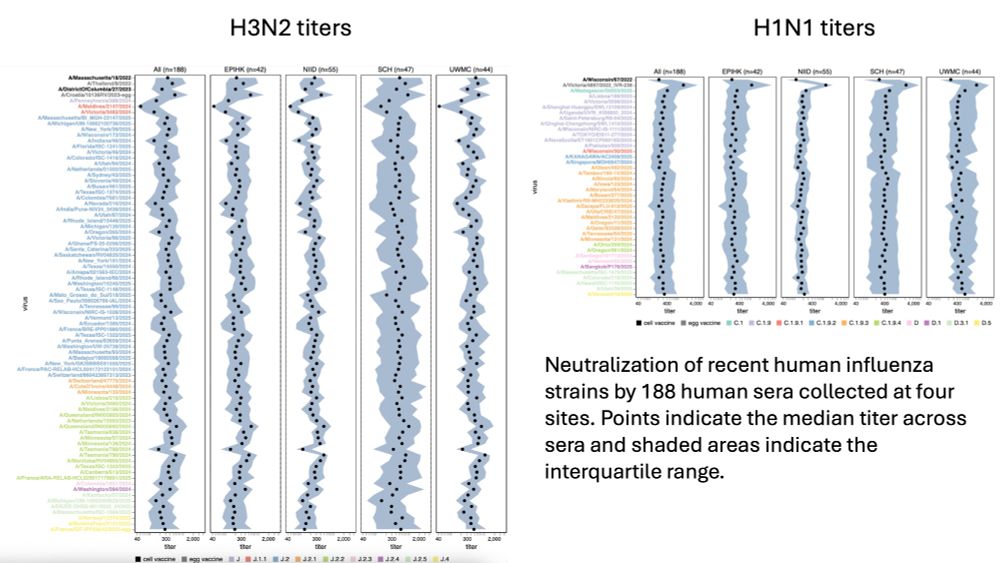
Titers are summarized below; can be examined interactively at jbloomlab.github.io/flu-seqneut-... & jbloomlab.github.io/flu-seqneut-...

Approach enabled one grad student (@ckikawa.bsky.social) to measure ~26,000 neutralization curves in ~5 months.

Approach enabled one grad student (@ckikawa.bsky.social) to measure ~26,000 neutralization curves in ~5 months.
Our goal was to use new approach to characterize human antibody landscape at scale in near real-time.

Our goal was to use new approach to characterize human antibody landscape at scale in near real-time.
Thanks to Xiaohui Ju for leading study
Special thanks to @msdiamondlab.bsky.social for help
Also Will Hannon, Caelan Radford, Brendan Larsen, Daved Fremont, Ofer Zimmerman, Tomasz Kaszuba, Chris Nelson, Israel Baltazar-Perez, Samantha Nelson

Thanks to Xiaohui Ju for leading study
Special thanks to @msdiamondlab.bsky.social for help
Also Will Hannon, Caelan Radford, Brendan Larsen, Daved Fremont, Ofer Zimmerman, Tomasz Kaszuba, Chris Nelson, Israel Baltazar-Perez, Samantha Nelson
So we reduced natural tropism for both human & mosquito cells to just one type of cell.

So we reduced natural tropism for both human & mosquito cells to just one type of cell.

We also find sites where mutations specifically impair entry in C6/36 cells. Although mosquito receptor unknown, we hypothesize these sites at its binding interface.

We also find sites where mutations specifically impair entry in C6/36 cells. Although mosquito receptor unknown, we hypothesize these sites at its binding interface.
For instance, mutations at E2 site 119 are generally tolerated in C6/36 and 293T-TIM1 cells, but deleterious in 293T-MXRA8 cells.
(See dms-vep.org/CHIKV-181-25... for interactive plot.)

For instance, mutations at E2 site 119 are generally tolerated in C6/36 and 293T-TIM1 cells, but deleterious in 293T-MXRA8 cells.
(See dms-vep.org/CHIKV-181-25... for interactive plot.)
Below is constraint mapped on structure; see dms-vep.org/CHIKV-181-25... for interactive heatmap of these data.

Below is constraint mapped on structure; see dms-vep.org/CHIKV-181-25... for interactive heatmap of these data.

A receptor for these viral proteins in mammalian cells is the protein MXRA8, but receptor in mosquito cells is unknown.

A receptor for these viral proteins in mammalian cells is the protein MXRA8, but receptor in mosquito cells is unknown.
Infection can cause fever and severe joint pain in humans.
Outbreaks are growing due to expanding mosquito range: www.nytimes.com/2025/08/19/h...

Infection can cause fever and severe joint pain in humans.
Outbreaks are growing due to expanding mosquito range: www.nytimes.com/2025/08/19/h...

Many of these sites have mutated during SARS-CoV-2 evolution in humans, demonstrating importance of RBD motion and its effects on ACE2 binding & antibody neutralization.

Many of these sites have mutated during SARS-CoV-2 evolution in humans, demonstrating importance of RBD motion and its effects on ACE2 binding & antibody neutralization.


So new exposures are altering neutralizing serum antibody repertoire, although our data do not define mechanism (see preprint for hypotheses).

So new exposures are altering neutralizing serum antibody repertoire, although our data do not define mechanism (see preprint for hypotheses).


A number of mutations that have spread recently are more tolerated in KP.3.1.1 than older XBB.1.5 variant, suggesting epistatic shifts favored their emergence.

A number of mutations that have spread recently are more tolerated in KP.3.1.1 than older XBB.1.5 variant, suggesting epistatic shifts favored their emergence.
Here we used approach to measure how mutations to KP.3.1.1 spike affect five phenotypes, as shown below.

Here we used approach to measure how mutations to KP.3.1.1 spike affect five phenotypes, as shown below.
KP.3.1.1 & other recent variants have >60 spike amino-acid mutations relative to early pandemic strains, as spike has evolved at extraordinary rate of >10 mutations/year on average.

KP.3.1.1 & other recent variants have >60 spike amino-acid mutations relative to early pandemic strains, as spike has evolved at extraordinary rate of >10 mutations/year on average.
Stabilization directed more neutralization towards potently neutralizing epitopes on top of HA head as shown below.

Stabilization directed more neutralization towards potently neutralizing epitopes on top of HA head as shown below.

