Posts
Media
Videos
Starter Packs
Reposted by Jerome
Reposted by Jerome
Jingyou Rao
@jingyour.bsky.social
· Aug 4

Cosmos: A Position-Resolution Causal Model for Direct and Indirect Effects in Protein Functions
Multi-phenotype deep mutational scanning (DMS) experiments provide a powerful means to dissect how protein variants affect different layers of molecular function, such as abundance, surface expression...
www.biorxiv.org
Jerome
@jeromics.bsky.social
· Jun 30
Jerome
@jeromics.bsky.social
· Jun 30

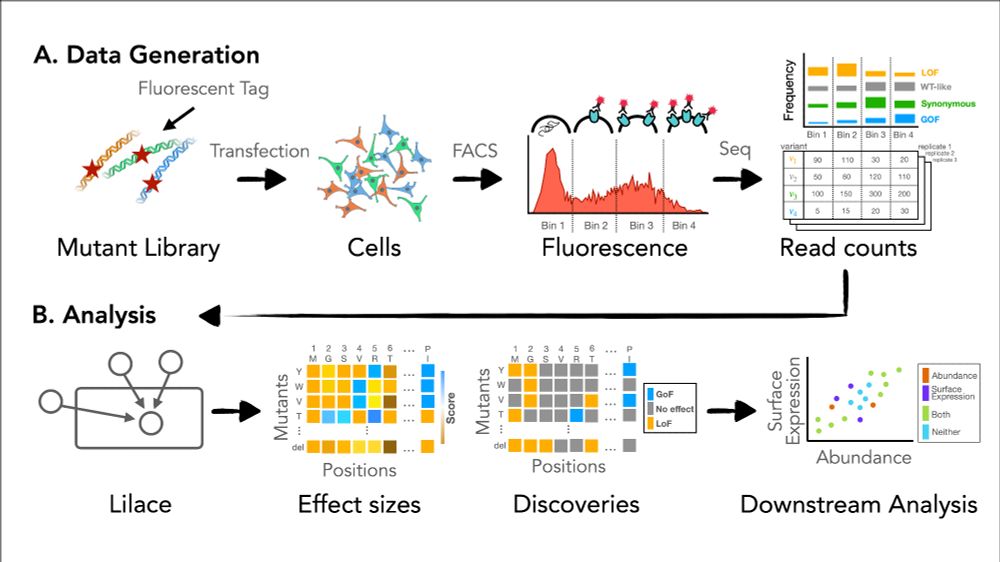
Accurate variant effect estimation in FACS-based deep mutational scanning data with Lilace
Deep mutational scanning (DMS) experiments interrogate the effect of genetic variants on protein function, often using fluorescence-activated cell sorting (FACS) to quantitatively measure molecular ph...
www.biorxiv.org