Jingyou Rao
@jingyour.bsky.social
150 followers
130 following
25 posts
Incoming Postdoc @UCSF wcoyotelab.com | PhD in Computer Science @UCLA protein epistasis and mutational scanning
Posts
Media
Videos
Starter Packs
Reposted by Jingyou Rao
Reposted by Jingyou Rao
Reposted by Jingyou Rao
Reposted by Jingyou Rao
Reposted by Jingyou Rao
Reposted by Jingyou Rao
Harold Pimentel
@hjp.bsky.social
· Aug 15
Jingyou Rao
@jingyour.bsky.social
· Aug 4

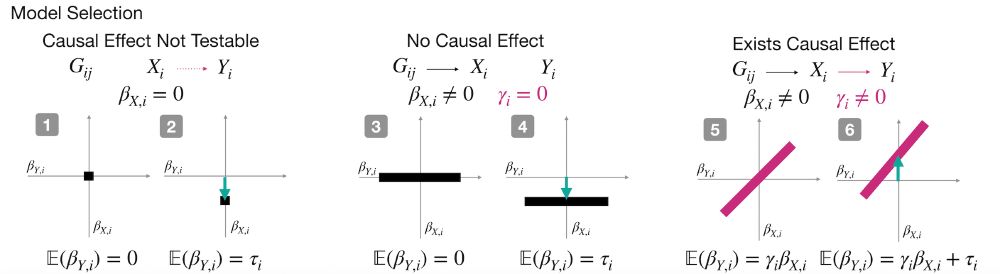
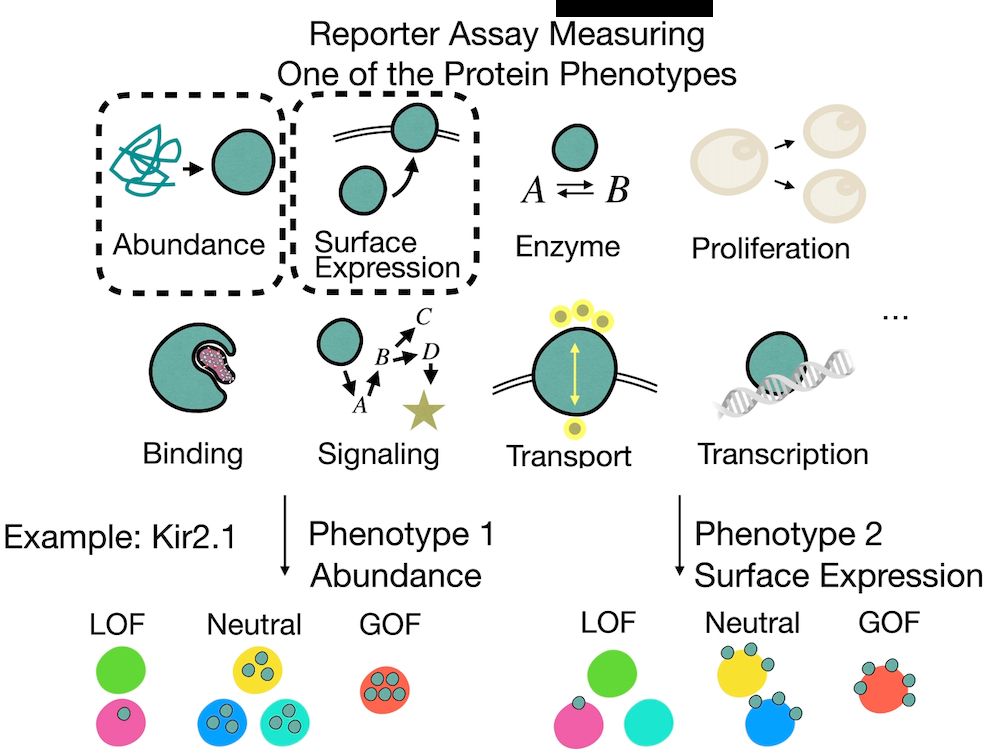
Cosmos: A Position-Resolution Causal Model for Direct and Indirect Effects in Protein Functions
Multi-phenotype deep mutational scanning (DMS) experiments provide a powerful means to dissect how protein variants affect different layers of molecular function, such as abundance, surface expression...
www.biorxiv.org
Reposted by Jingyou Rao
Roshni Patel
@roshnipatel.bsky.social
· Aug 6
Jingyou Rao
@jingyour.bsky.social
· Aug 4
Jingyou Rao
@jingyour.bsky.social
· Aug 4

Cosmos: A Position-Resolution Causal Model for Direct and Indirect Effects in Protein Functions
Multi-phenotype deep mutational scanning (DMS) experiments provide a powerful means to dissect how protein variants affect different layers of molecular function, such as abundance, surface expression...
www.biorxiv.org
Reposted by Jingyou Rao
Reposted by Jingyou Rao
Reposted by Jingyou Rao
Doug Fowler
@dougfowler.bsky.social
· Jul 21
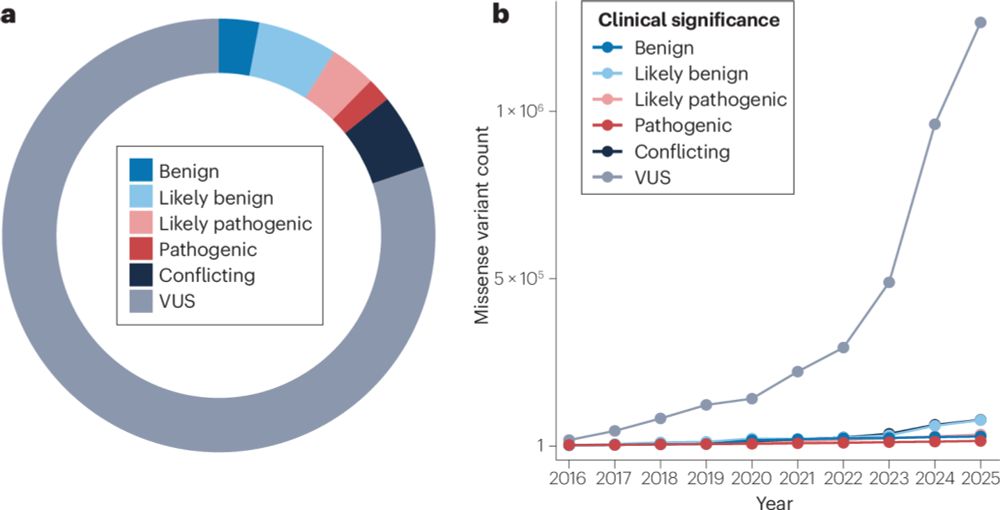
Multiplexed assays of variant effect for clinical variant interpretation
Nature Reviews Genetics - Multiplexed assays of variant effect (MAVEs) are highly scalable experimental approaches used to generate functional data for genetic variants. In this Review, McEwen et...
www.nature.com
Reposted by Jingyou Rao
Reposted by Jingyou Rao
Jerome
@jeromics.bsky.social
· Jun 30

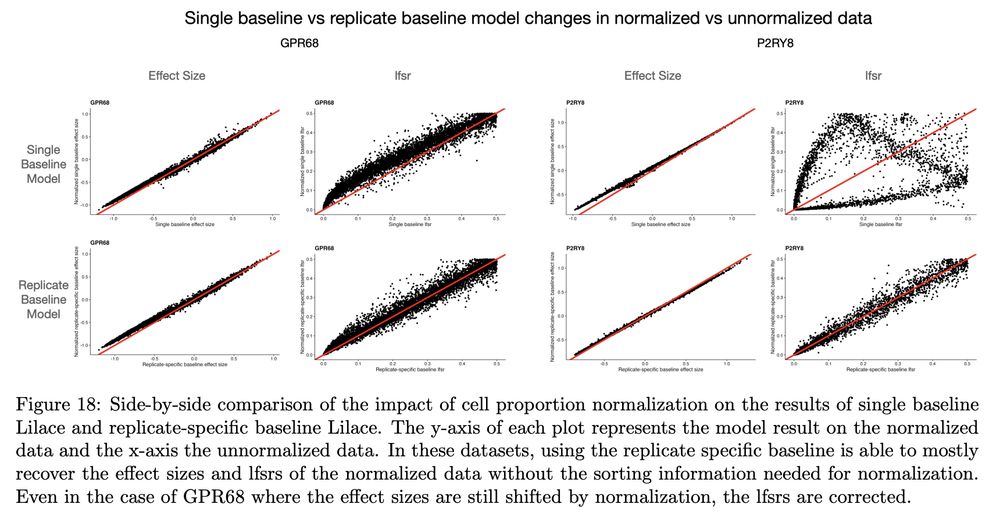
Accurate variant effect estimation in FACS-based deep mutational scanning data with Lilace
Deep mutational scanning (DMS) experiments interrogate the effect of genetic variants on protein function, often using fluorescence-activated cell sorting (FACS) to quantitatively measure molecular ph...
www.biorxiv.org
Reposted by Jingyou Rao