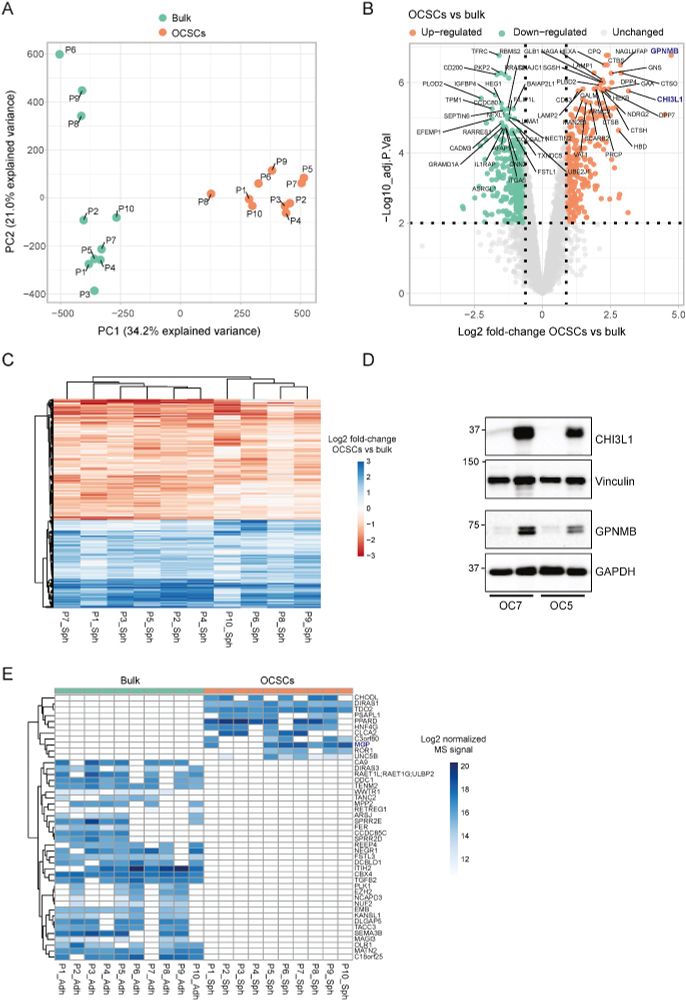
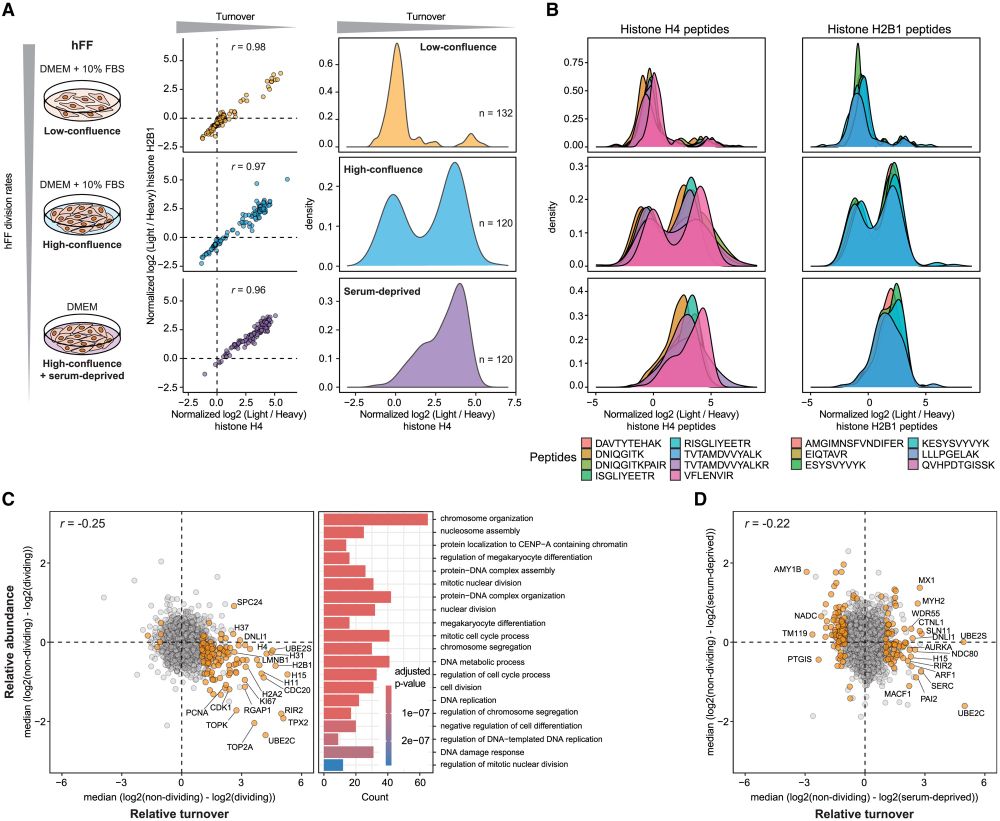
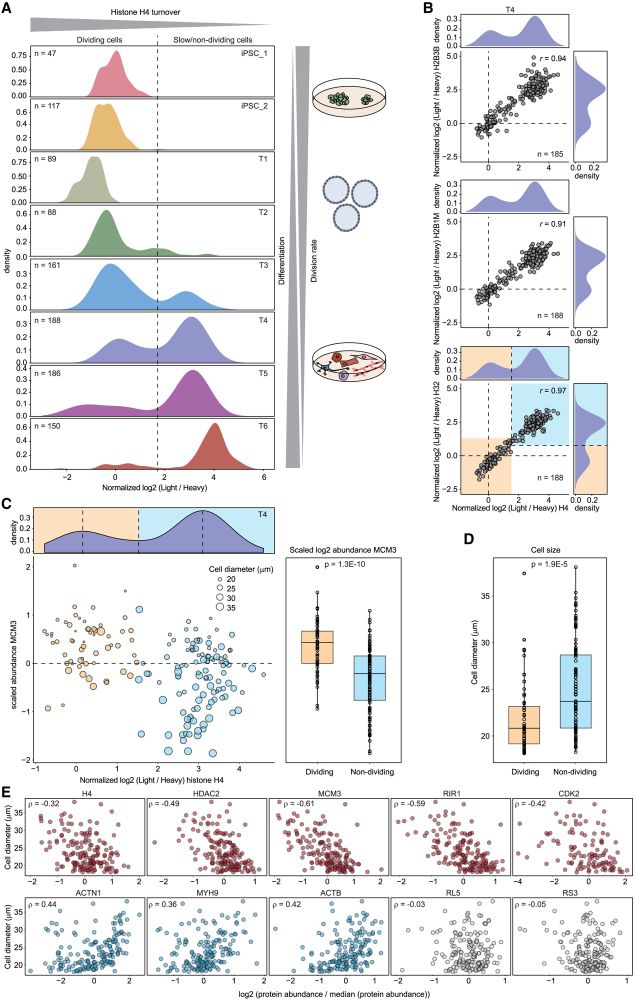
JesperOlsenLab
@jesperolsenlab.bsky.social
630 followers
90 following
36 posts
Mass spectrometry for quantitative proteomics. The Olsen group at the Center for Protein Research, University of Copenhagen.
Posts
Media
Videos
Starter Packs
Palesa
@palaeoprotpalesa.bsky.social
· May 29
Enamel proteins reveal biological sex and genetic variability in southern African Paranthropus
Paranthropus robustus is a morphologically well-documented Early Pleistocene hominin species from southern Africa with no genetic evidence reported so far. In this work, we describe the mass spectrome...
doi.org