
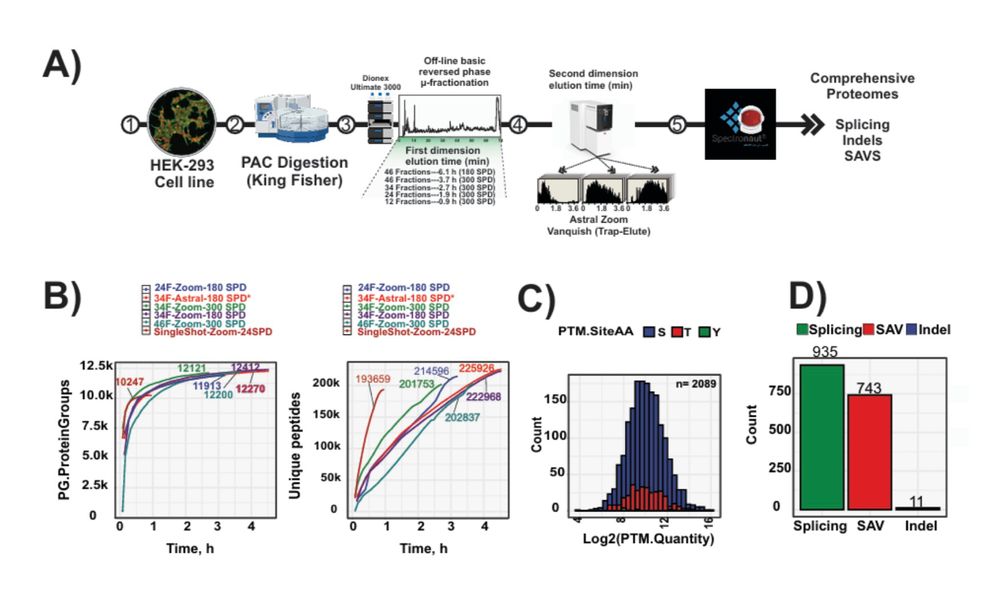
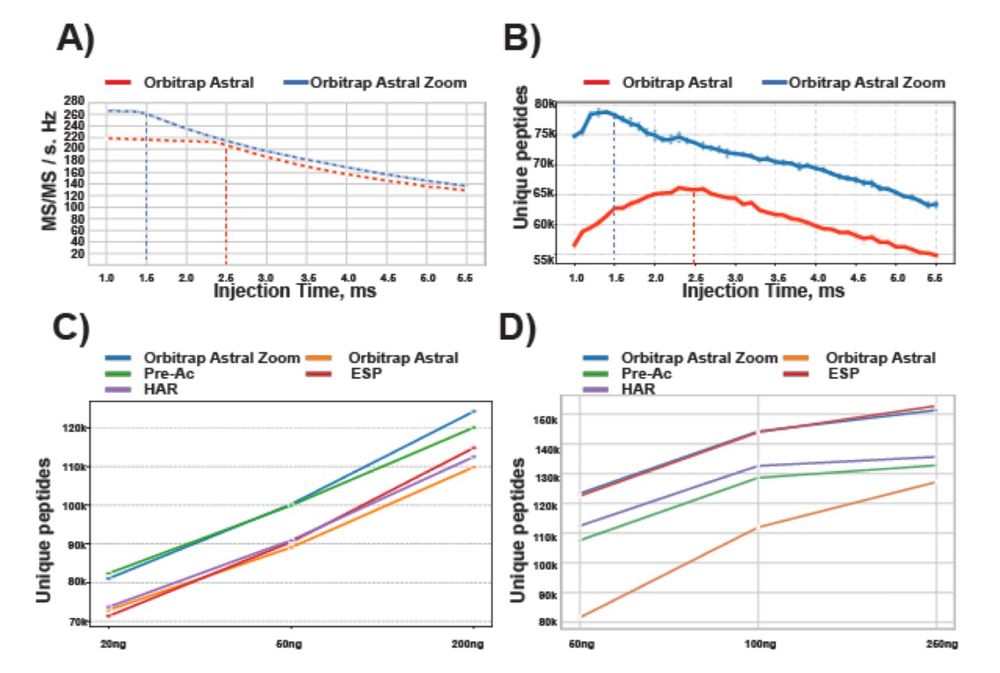
The benefits are even higher with ΦSDM, especially in fast, low-resolution Orbitrap runs.
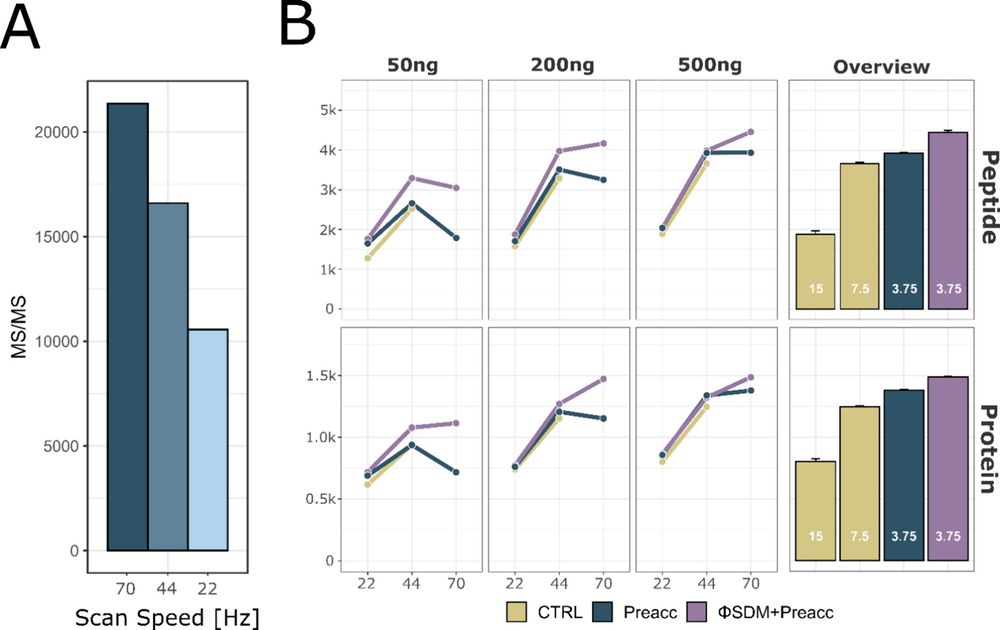
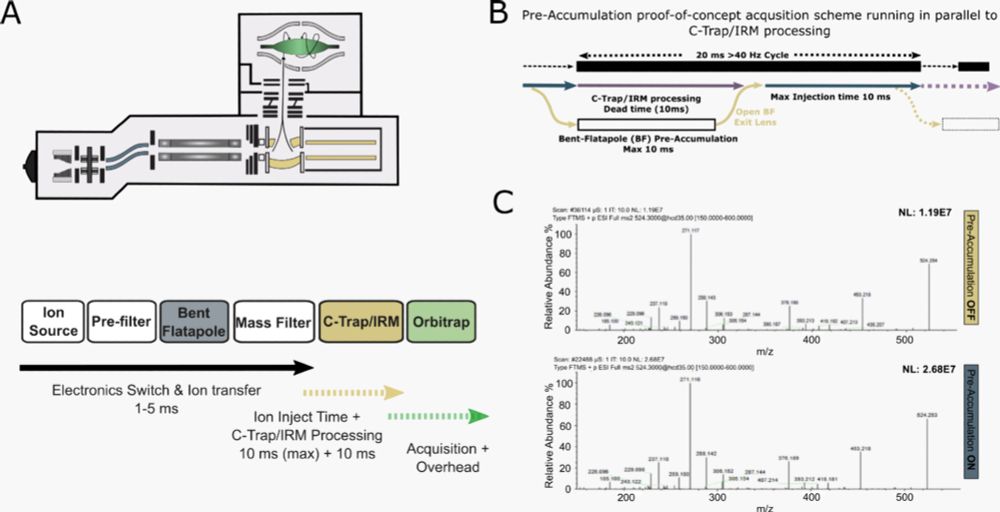
The benefits are even higher with ΦSDM, especially in fast, low-resolution Orbitrap runs.
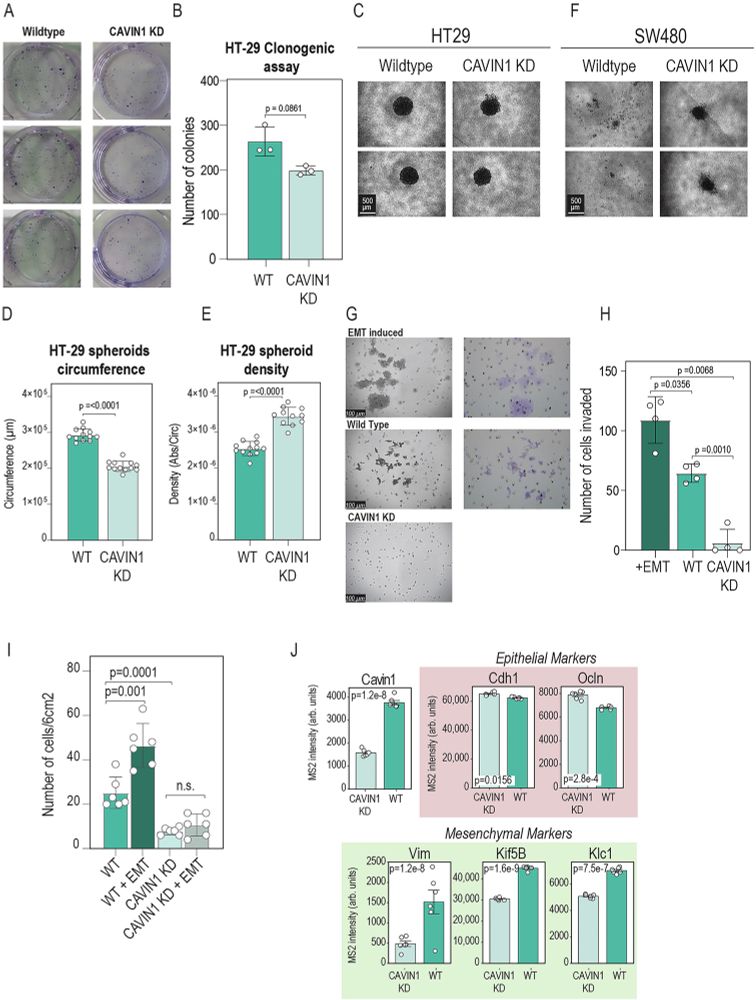
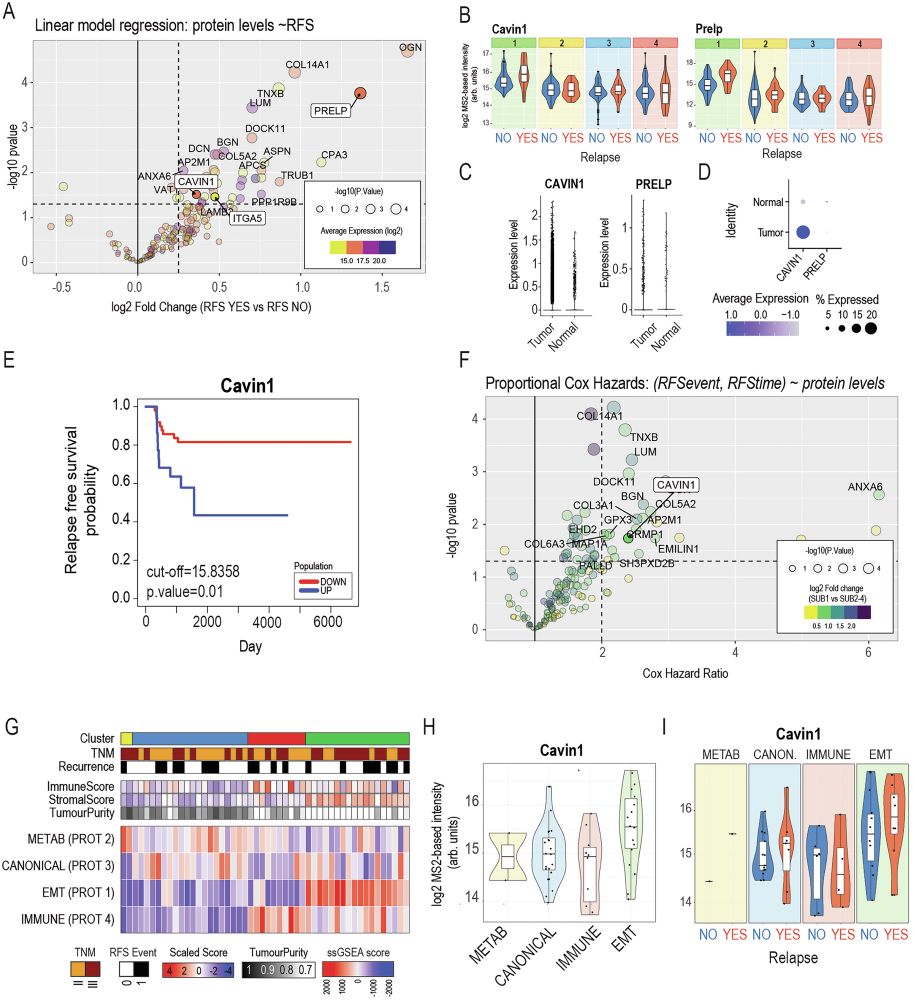
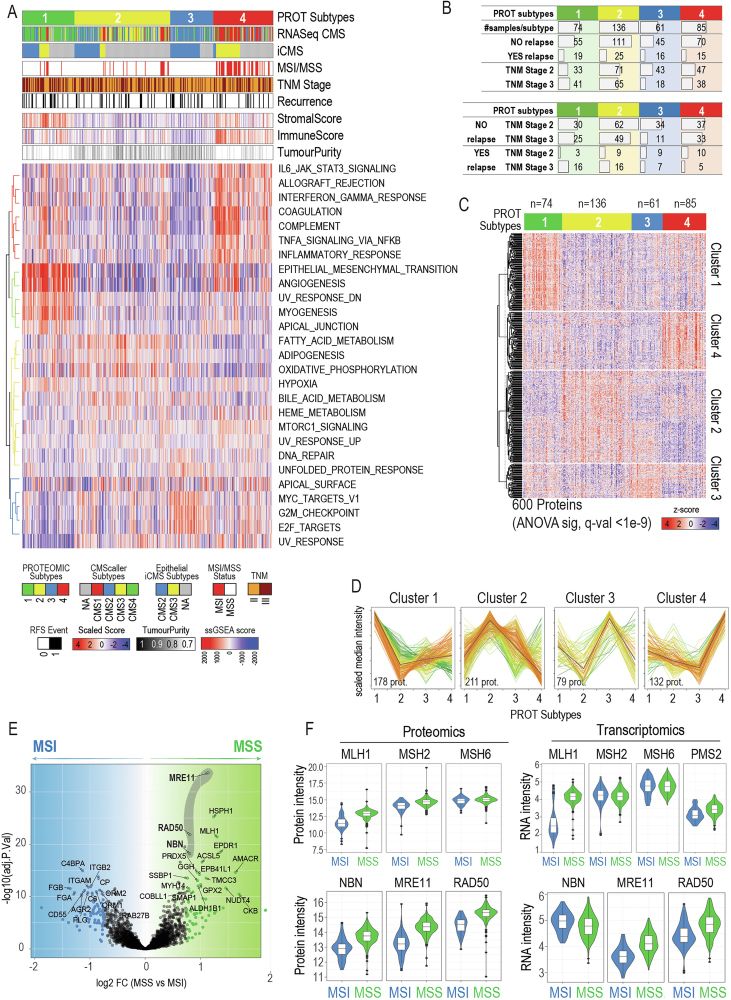
When we treated patient-derived ovarian cancer cells with a PDGFR inhibitor, their stemness potential dropped (sphere formation ↓).
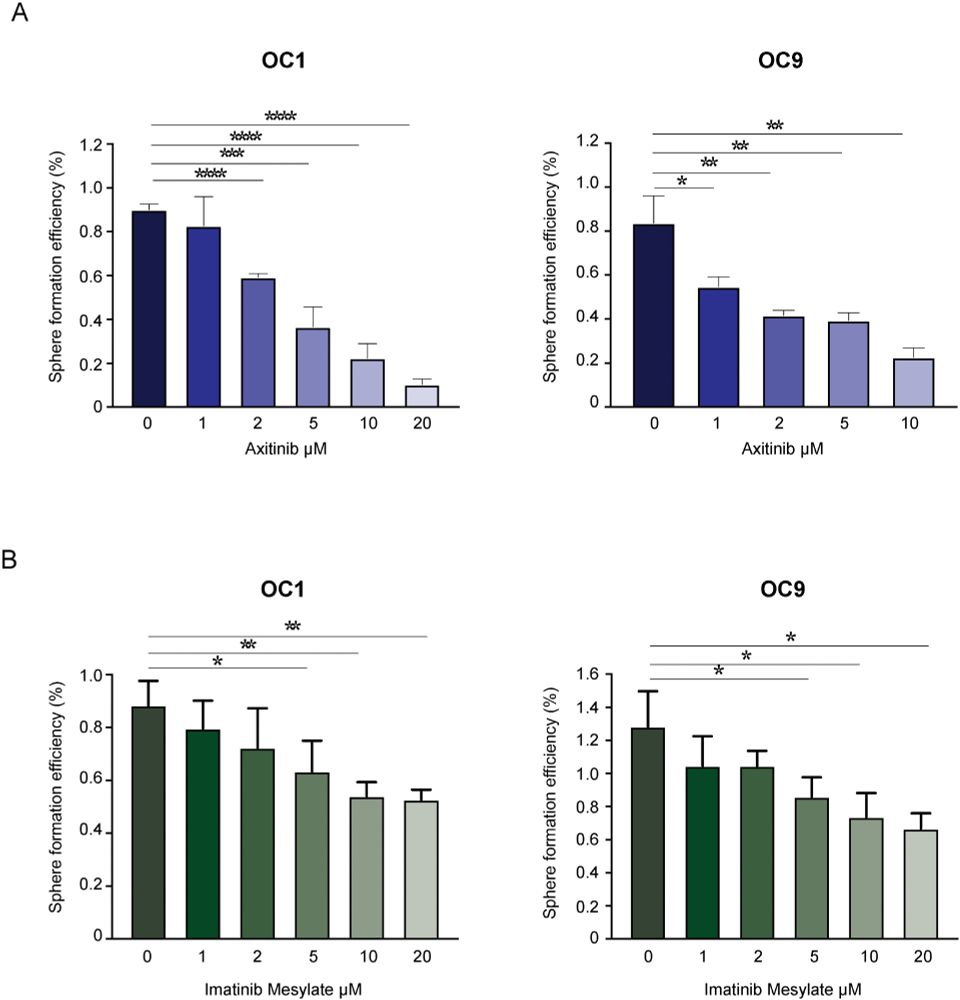
When we treated patient-derived ovarian cancer cells with a PDGFR inhibitor, their stemness potential dropped (sphere formation ↓).
That’s huge: it means this signature might help predict therapy resistance.
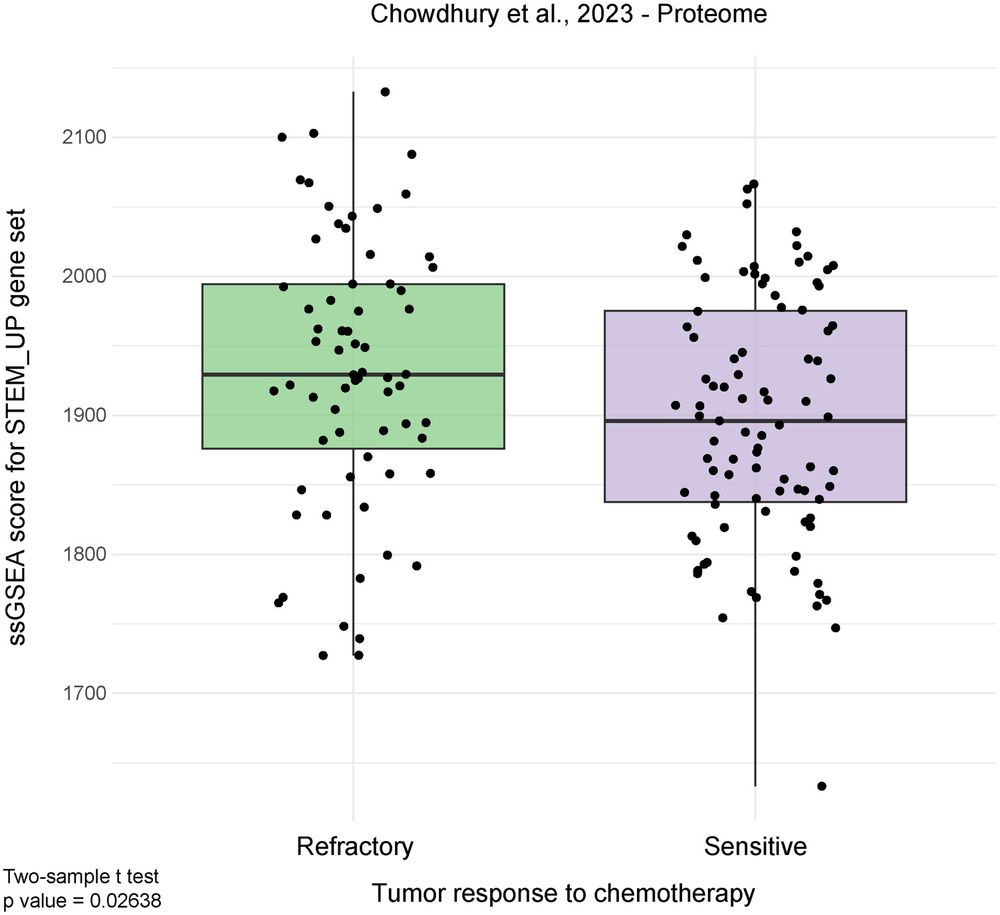
That’s huge: it means this signature might help predict therapy resistance.
This points to a link between the stem-like state and the tumor microenvironment.
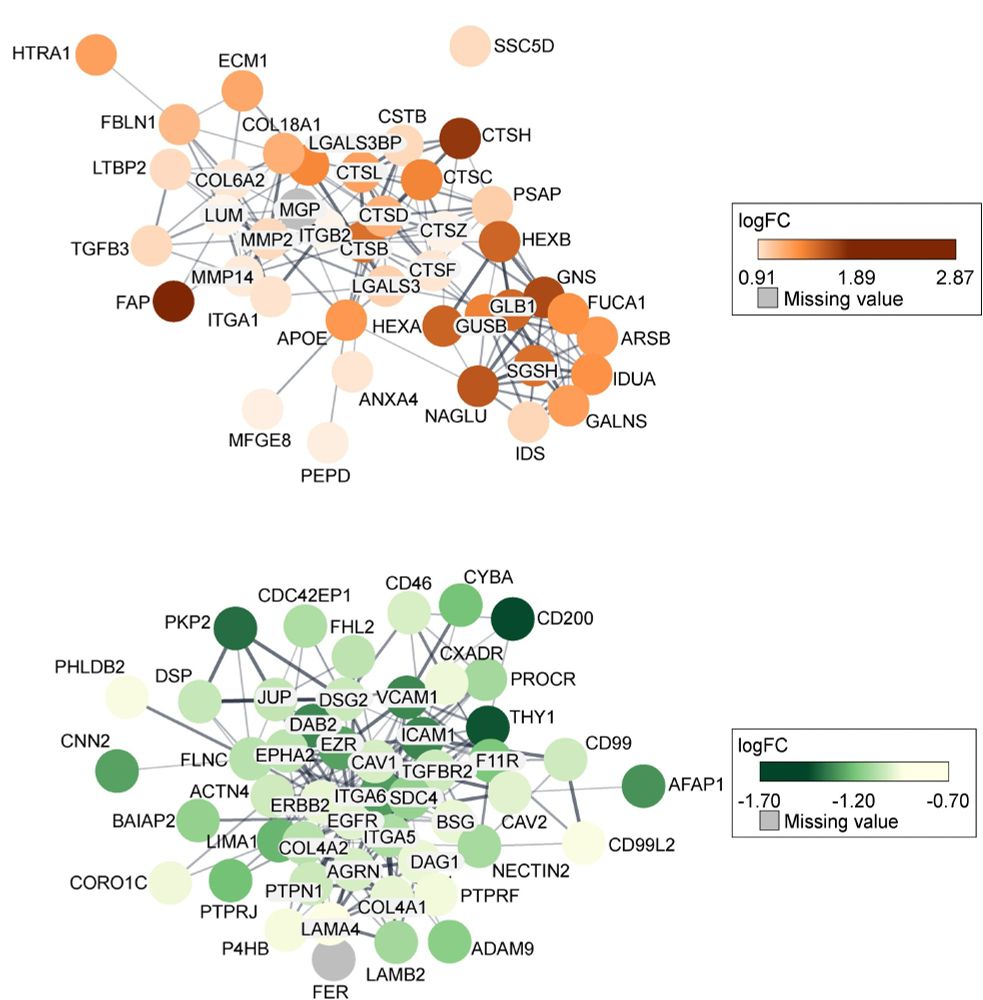
This points to a link between the stem-like state and the tumor microenvironment.
This revealed a shared stemness-related signature across patients, despite high inter-patient heterogeneity.
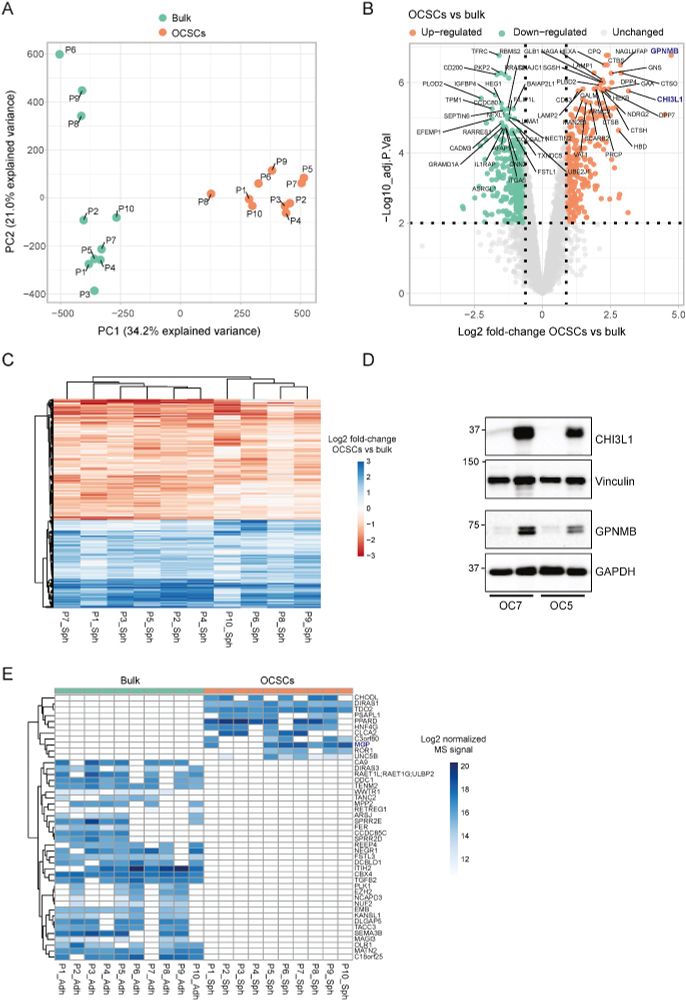
This revealed a shared stemness-related signature across patients, despite high inter-patient heterogeneity.
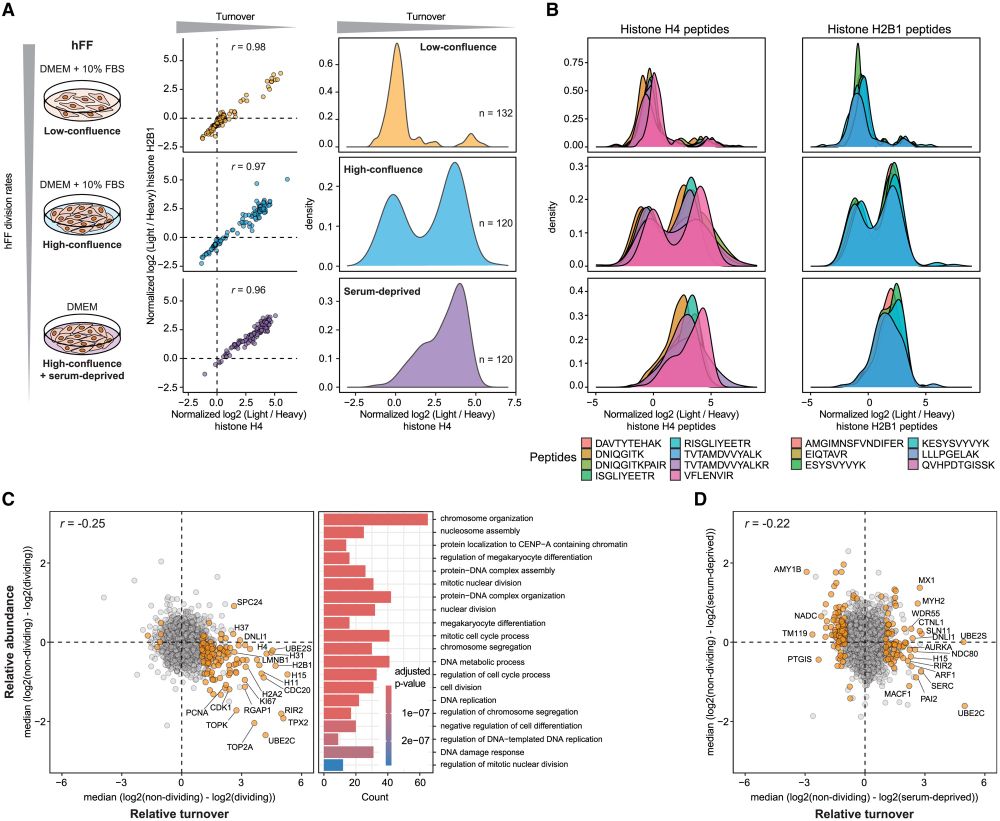
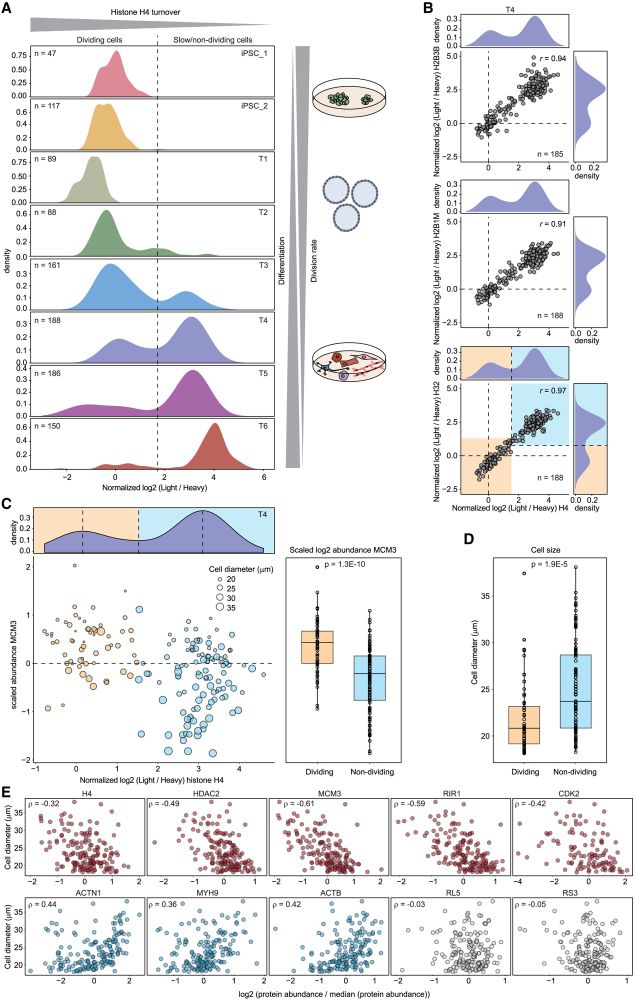
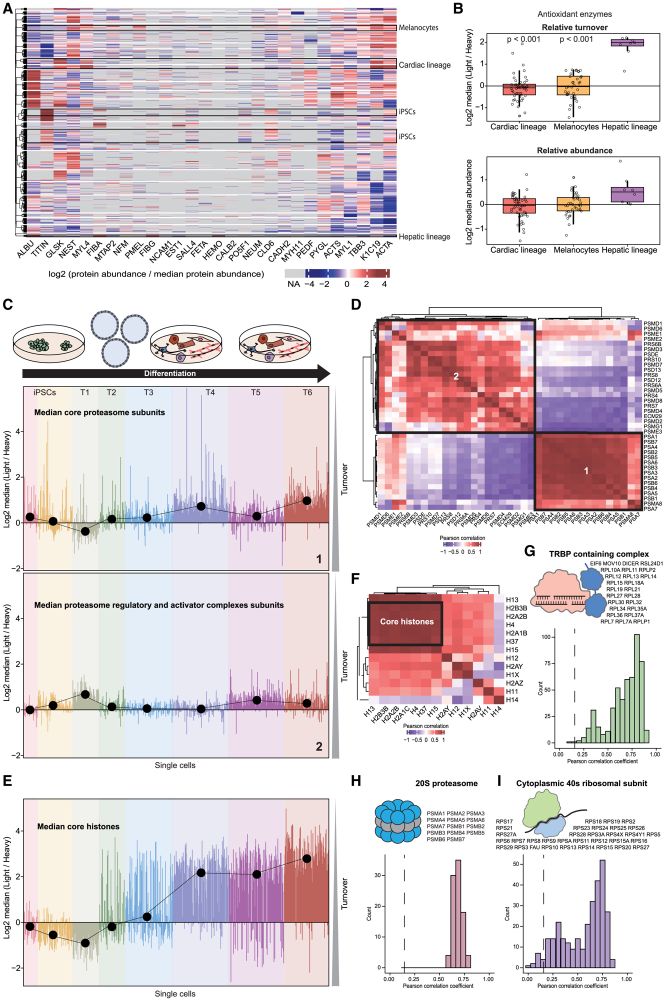
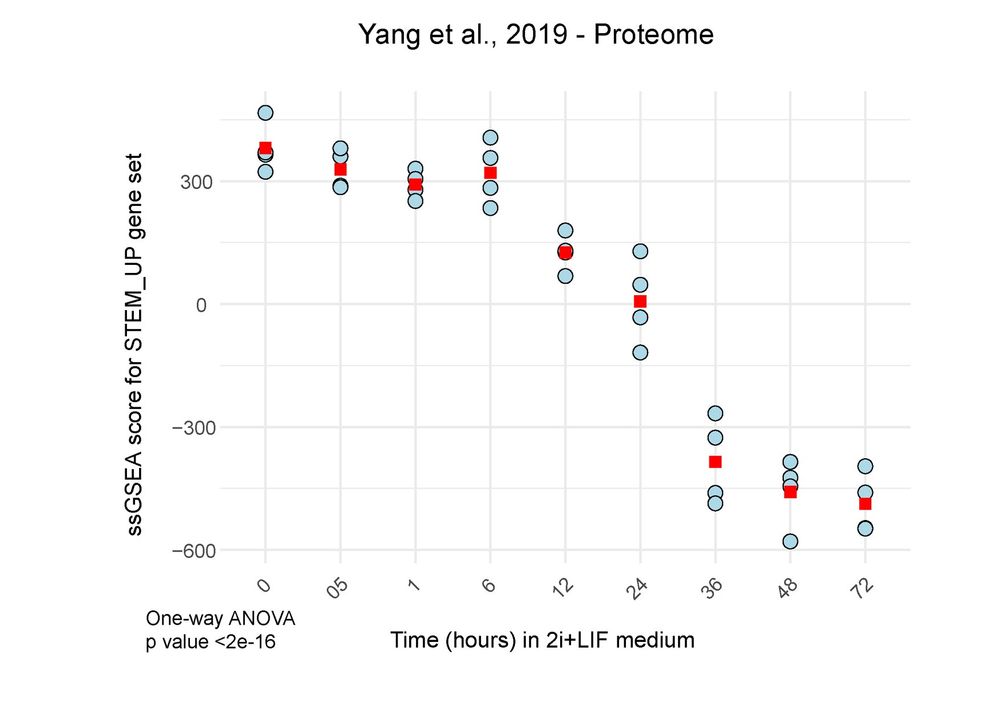
Our comprehensive dataset shows protein turnover dynamics during cell differentiation over 2 months.
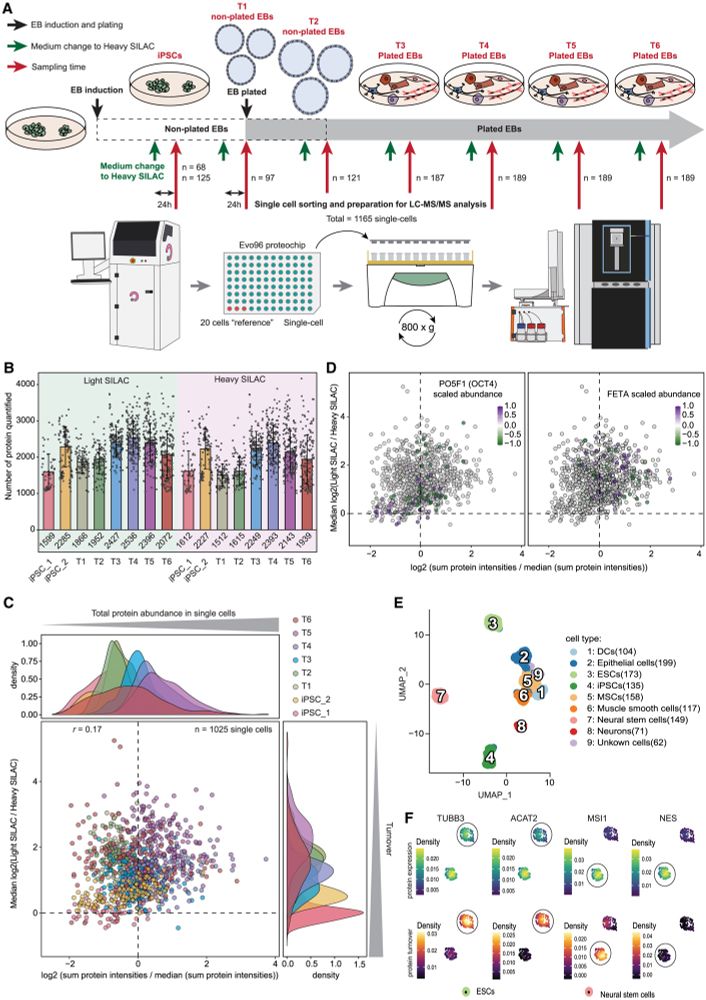
Our comprehensive dataset shows protein turnover dynamics during cell differentiation over 2 months.
SC-pSILAC revealed treatment-related changes in single cells.
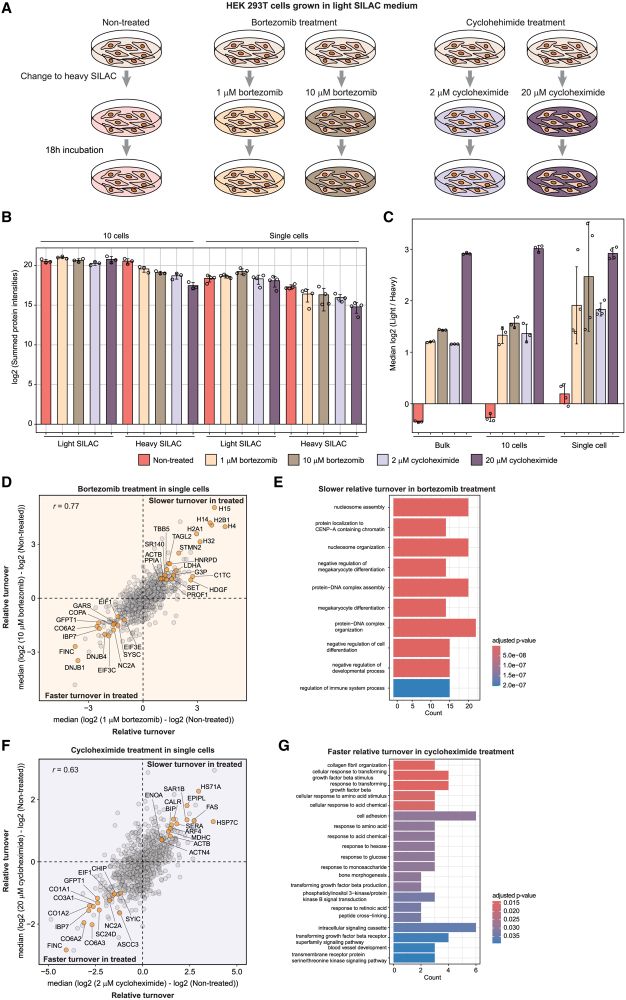
SC-pSILAC revealed treatment-related changes in single cells.

