Joe Greener
@jgreener64.bsky.social
530 followers
420 following
140 posts
Computational chemist/structural bioinformatician working on improving molecular simulation at MRC Laboratory of Molecular Biology. jgreener64.github.io
Posts
Media
Videos
Starter Packs
Pinned
Joe Greener
@jgreener64.bsky.social
· May 28
Reposted by Joe Greener
Carl T. Bergstrom
@carlbergstrom.com
· Jul 16

Will anyone review this paper? Screening, sorting, and the feedback cycles that imperil peer review
Scholarly publishing relies on peer review to identify the best science. Yet finding willing and qualified reviewers to evaluate manuscripts has become an increasingly challenging task, possibly even ...
arxiv.org
Reposted by Joe Greener
Reposted by Joe Greener
Reposted by Joe Greener
COSMO Lab
@labcosmo.bsky.social
· Jun 20
The dark side of the forces: assessing non-conservative force...
The use of machine learning to estimate the energy of a group of atoms, and the forces that drive them to more stable configurations, have revolutionized the fields of computational chemistry and...
openreview.net
Joe Greener
@jgreener64.bsky.social
· Jun 16

Postdoctoral Scientist - Structural Studies - Dr Joe Greener - LMB 2633 - Cambridgeshire job with MRC Laboratory of Molecular Biology (LMB) | 12840475
Postdoctoral Scientist Salary £41,344 per annum Fixed Term – 3 years MRC Laboratory of Molecular Biology, Cambridge, UK We are looking for a postd...
www.nature.com
Joe Greener
@jgreener64.bsky.social
· Jun 13
Reposted by Joe Greener
Reposted by Joe Greener
Philip Ball
@philipcball.bsky.social
· May 31
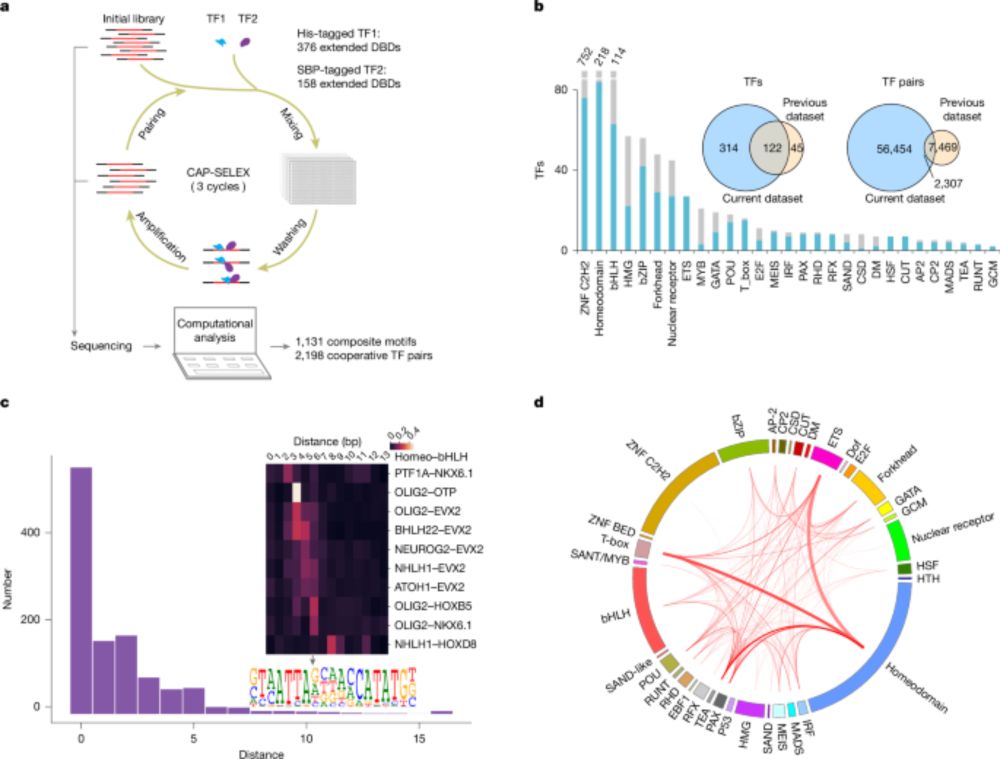
DNA-guided transcription factor interactions extend human gene regulatory code - Nature
A large-scale analysis of DNA-bound transcription factors (TFs) shows how the presence of DNA markedly affects the landscape of TF interactions, and identifies composite motifs that are recognized by ...
www.nature.com
Joe Greener
@jgreener64.bsky.social
· May 28
Reposted by Joe Greener
Sjors Scheres
@sjorsscheres.bsky.social
· May 28

Cryo-EM structure of the vault from human brain reveals symmetry mismatch at its caps
The vault protein is expressed in most eukaryotic cells, where it is assembled on polyribosomes into large hollow barrel-shaped complexes. Despite its widespread and abundant presence in cells, the bi...
www.biorxiv.org
Joe Greener
@jgreener64.bsky.social
· May 19
Cole Group
@colegroupncl.bsky.social
· May 19

MACE-OFF: Short-Range Transferable Machine Learning Force Fields for Organic Molecules
Classical empirical force fields have dominated biomolecular simulations for over 50 years. Although widely used in drug discovery, crystal structure prediction, and biomolecular dynamics, they generally lack the accuracy and transferability required for first-principles predictive modeling. In this paper, we introduce MACE-OFF, a series of short-range transferable force fields for organic molecules created using state-of-the-art machine learning technology and first-principles reference data computed with a high level of quantum mechanical theory. MACE-OFF demonstrates the remarkable capabilities of short-range models by accurately predicting a wide variety of gas- and condensed-phase properties of molecular systems. It produces accurate, easy-to-converge dihedral torsion scans of unseen molecules as well as reliable descriptions of molecular crystals and liquids, including quantum nuclear effects. We further demonstrate the capabilities of MACE-OFF by determining free energy surfaces in explicit solvent as well as the folding dynamics of peptides and nanosecond simulations of a fully solvated protein. These developments enable first-principles simulations of molecular systems for the broader chemistry community at high accuracy and relatively low computational cost.
pubs.acs.org
Joe Greener
@jgreener64.bsky.social
· May 16
Open Free Energy
@openfree.energy
· May 15
The Free Energy of Everything: Benchmarking OpenFE
Written by: Josh Horton, PhD
Who is OpenFE and what do they do?
Open Free Energy (OpenFE) sits at the nexus of academia and industry. We are developing an open-source software ecosystem for alchem...
blog.omsf.io
Joe Greener
@jgreener64.bsky.social
· May 9
Joe Greener
@jgreener64.bsky.social
· Apr 29