Joao Ascensao
@joaoascensao.bsky.social
490 followers
320 following
25 posts
Postdoc @ Harvard with Michael Desai | Evolutionary dynamics
ocf.berkeley.edu/~joaoascensao
Posts
Media
Videos
Starter Packs
Pinned
Joao Ascensao
@joaoascensao.bsky.social
· Jul 21
Reposted by Joao Ascensao
Joao Ascensao
@joaoascensao.bsky.social
· Aug 22
Joao Ascensao
@joaoascensao.bsky.social
· Aug 21

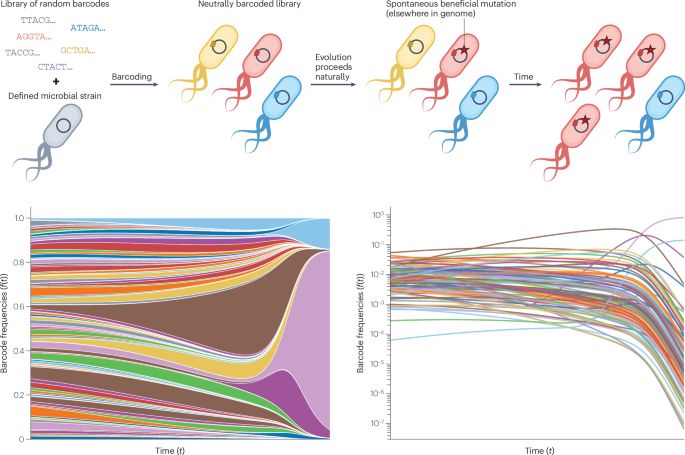
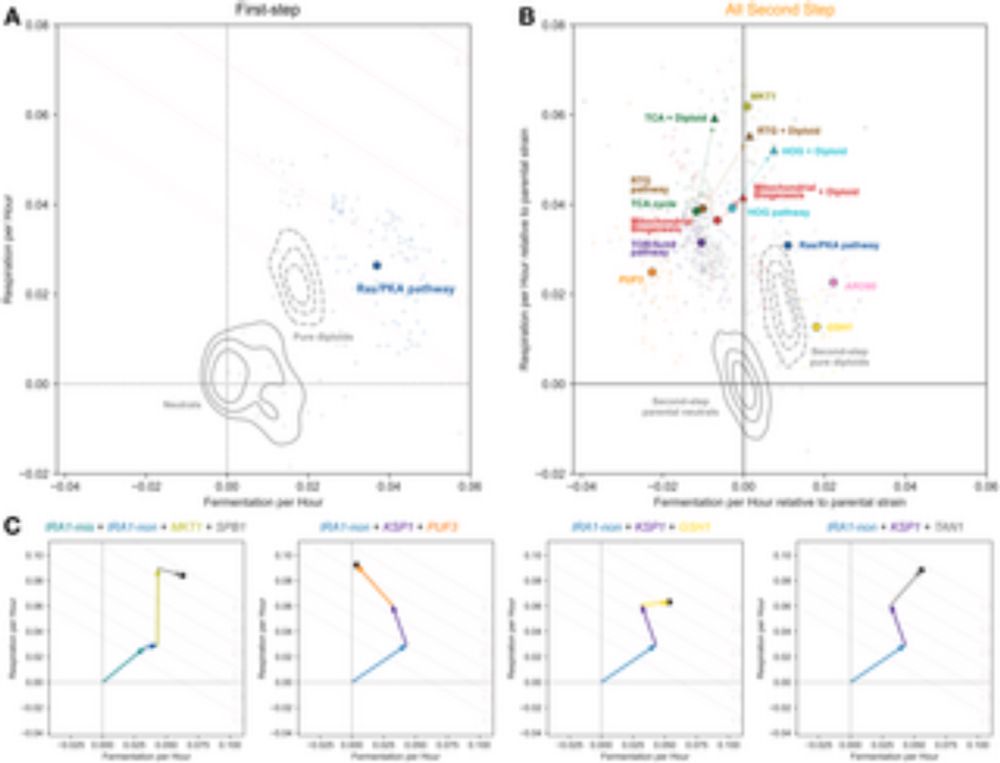
Frequency-dependent fitness effects are ubiquitous
In simple microbial populations, the fitness effects of most selected mutations are generally taken to be constant, independent of genotype frequency. This assumption underpins predictions about evolutionary dynamics, epistatic interactions, and the maintenance of genetic diversity in populations. Here, we systematically test this assumption using beneficial mutations from early generations of the Escherichia coli Long-Term Evolution Experiment (LTEE). Using flow cytometry-based competition assays, we find that frequency-dependent fitness effects are the norm rather than the exception, occurring in approximately 80\% of strain pairs tested. Most competitions exhibit negative frequency-dependence, where fitness advantages decline as mutant frequency increases. Furthermore, we demonstrate that the strength of frequency-dependence is predictable from invasion fitness measurements, with invasion fitness explaining approximately half of the biological variation in frequency-dependent slopes. Additionally, we observe violations of fitness transitivity in several strain combinations, indicating that competitive relationships cannot always be predicted from fitness relative to a single reference strain alone. Through high-resolution measurements of within-growth cycle dynamics, we show that simple resource competition explains a substantial portion of the frequency-dependence: when faster-growing genotypes dominate populations, they deplete shared resources more rapidly, reducing the time available for fitness differences to accumulate. Our results demonstrate that even in a simple model system designed to minimize ecological complexity, subtle ecological interactions between closely related genotypes create frequency-dependent selection that can fundamentally alter evolutionary dynamics. ### Competing Interest Statement The authors have declared no competing interest.
doi.org
Reposted by Joao Ascensao
Joao Ascensao
@joaoascensao.bsky.social
· Jul 21
Reposted by Joao Ascensao
Manuel Razo
@mrazo.bsky.social
· May 15
Reposted by Joao Ascensao
Joao Ascensao
@joaoascensao.bsky.social
· Nov 22
Joao Ascensao
@joaoascensao.bsky.social
· Nov 22
Joao Ascensao
@joaoascensao.bsky.social
· Nov 22
Joao Ascensao
@joaoascensao.bsky.social
· Nov 22

Asynchronous abundance fluctuations can drive giant genotype frequency fluctuations
Nature Ecology & Evolution - Based on a combination of experiments and modelling, this study shows large stochastic fluctuations in genotype frequencies caused by intrinsic and extrinsic...
rdcu.be
Joao Ascensao
@joaoascensao.bsky.social
· Feb 28
Joao Ascensao
@joaoascensao.bsky.social
· Feb 28
Joao Ascensao
@joaoascensao.bsky.social
· Feb 28
Joao Ascensao
@joaoascensao.bsky.social
· Feb 28
Joao Ascensao
@joaoascensao.bsky.social
· Feb 28
Joao Ascensao
@joaoascensao.bsky.social
· Feb 28
Joao Ascensao
@joaoascensao.bsky.social
· Feb 28
Joao Ascensao
@joaoascensao.bsky.social
· Feb 28