JungmannLab
@jungmannlab.bsky.social
150 followers
21 following
26 posts
Our group at LMU Munich and @mpibiochem.bsky.social uses DNA nanotechnology to develop next-generation super-resolution microscopy techniques. #DNAPAINT
Posts
Media
Videos
Starter Packs
Reposted by JungmannLab
JungmannLab
@jungmannlab.bsky.social
· Jul 30
Reposted by JungmannLab
Nature Nanotechnology
@natnano.nature.com
· Jul 28
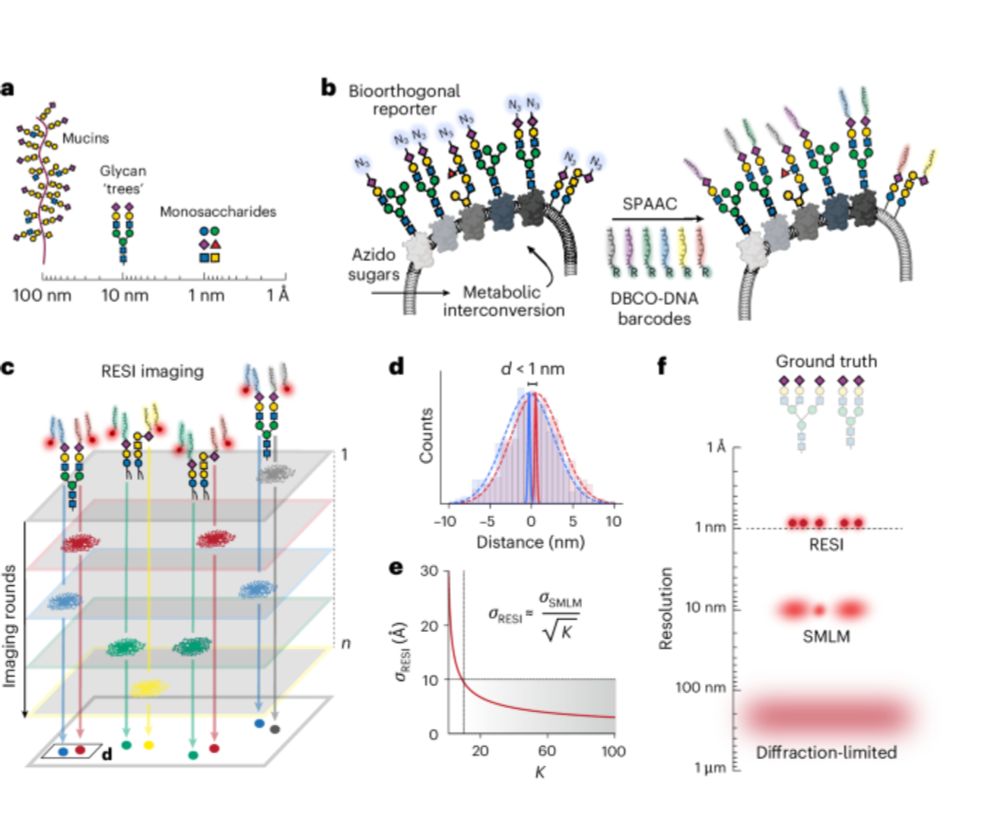
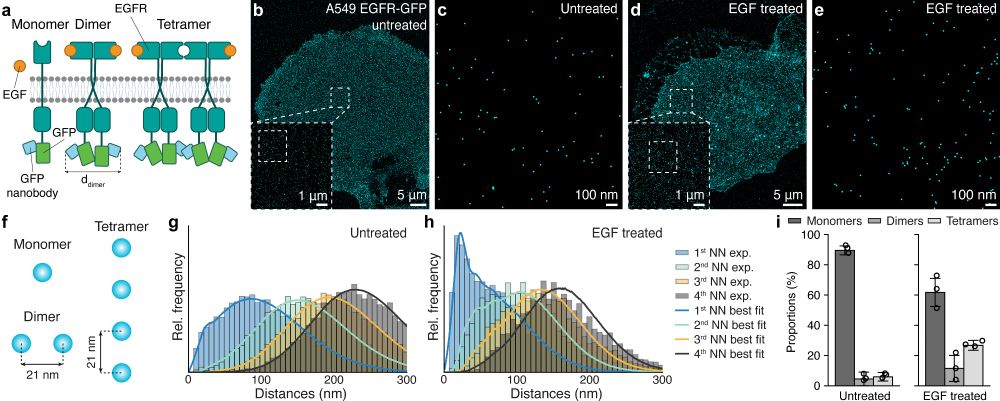
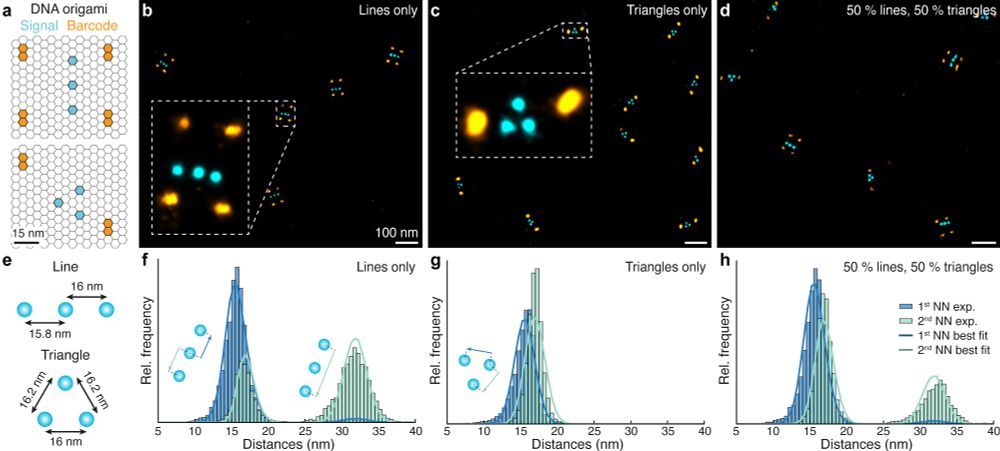
Ångström-resolution imaging of cell-surface glycans - Nature Nanotechnology
By combining bioorthogonal metabolic labelling and resolution enhancement through sequential imaging of DNA barcodes, the molecular organization of individual sugars in the native glycocalyx has been ...
www.nature.com
JungmannLab
@jungmannlab.bsky.social
· Jul 28
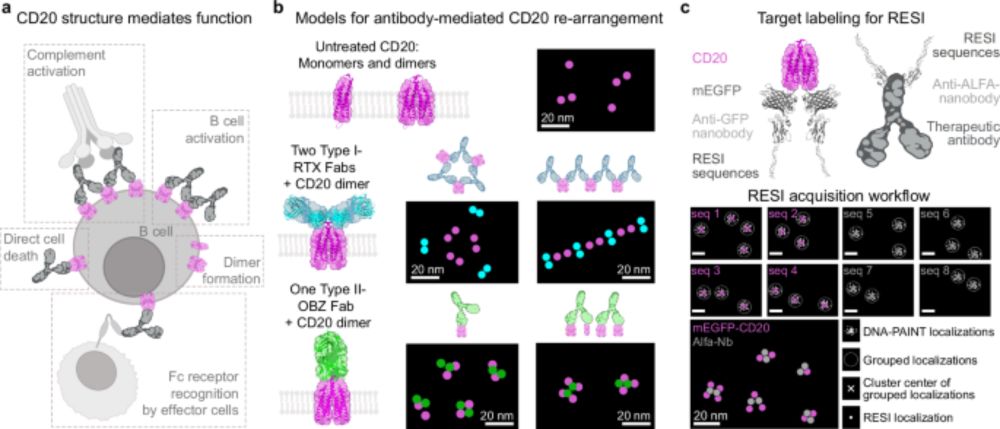
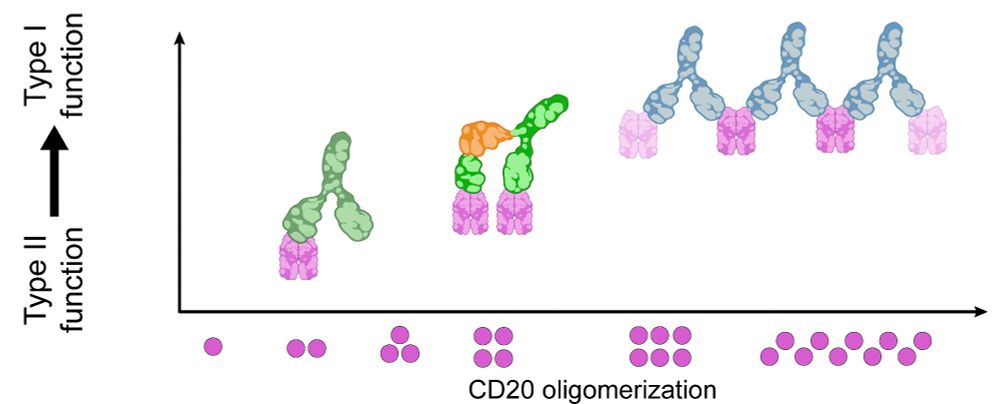
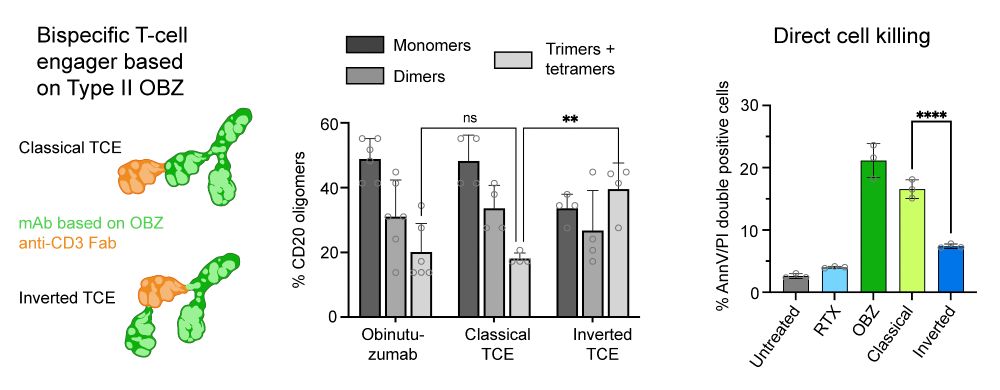
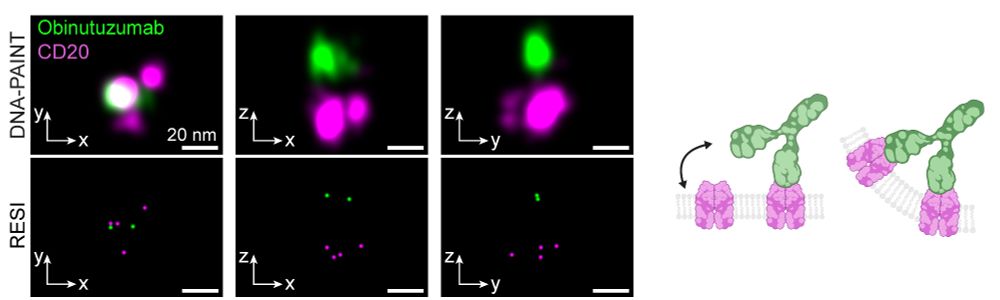
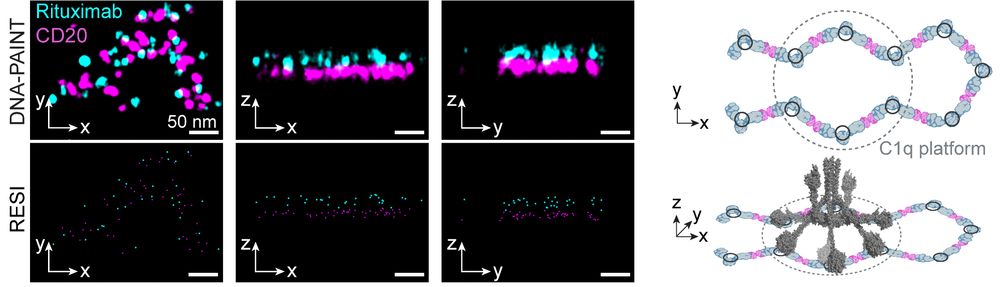
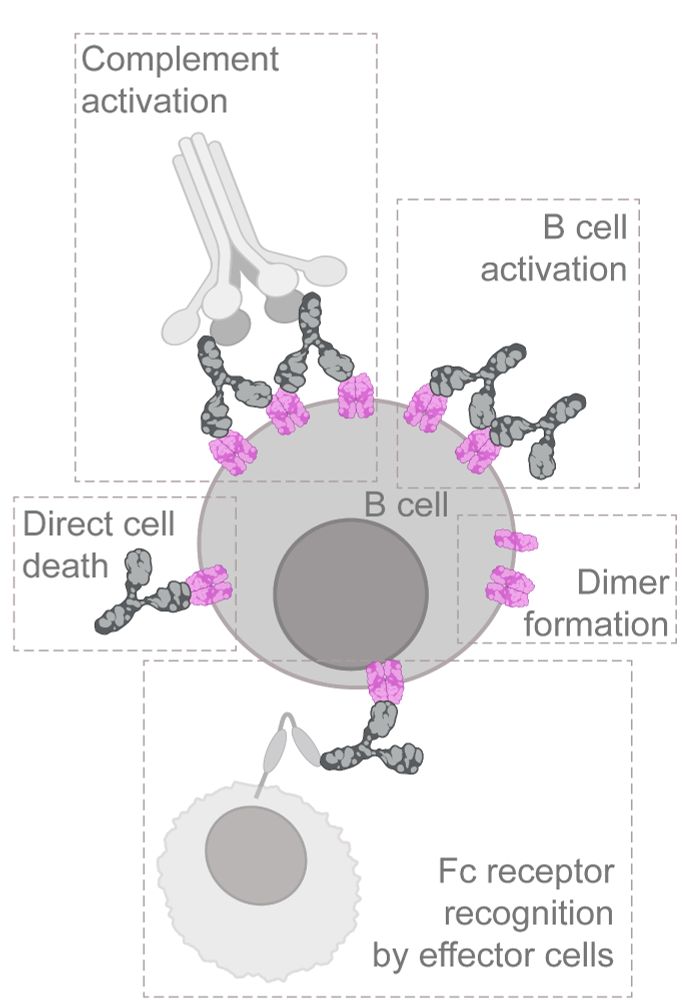
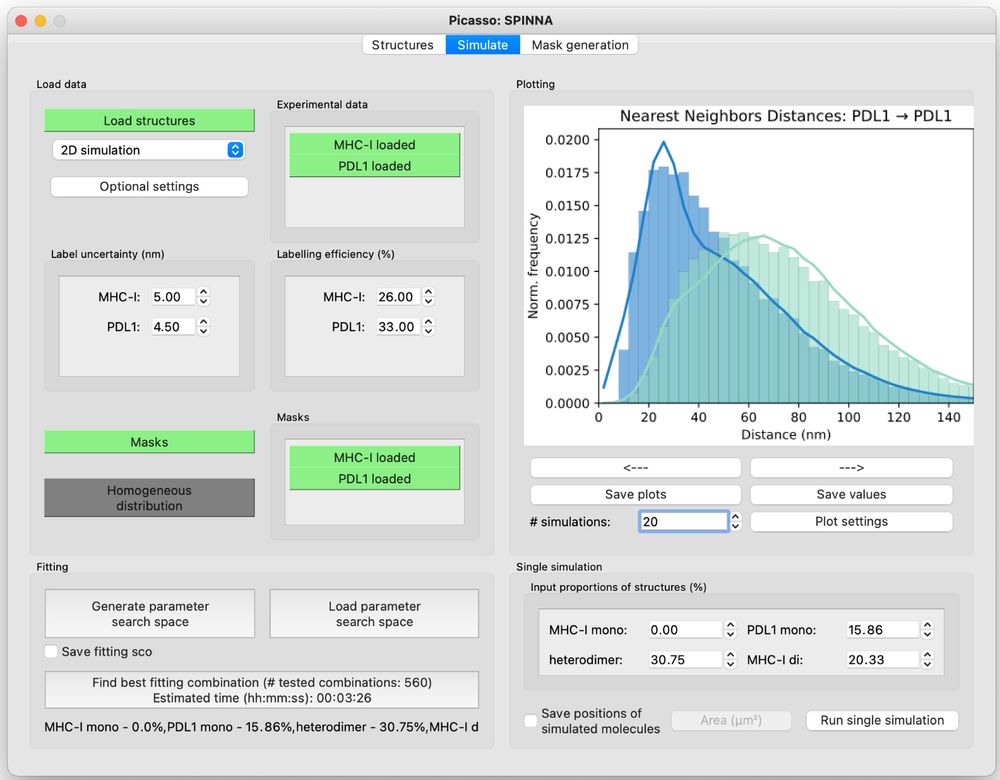
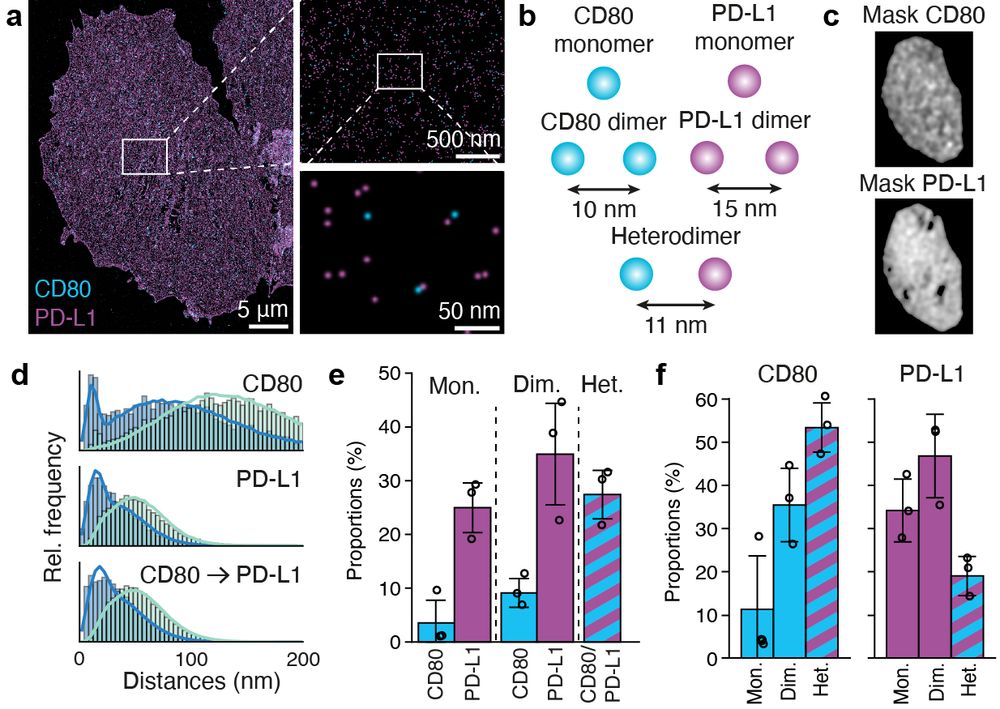
Resolving the structural basis of therapeutic antibody function in cancer immunotherapy with RESI - Nature Communications
The nanoscale organization of the antigen-antibody complexes influences the therapeutic action of monoclonal antibodies. Here, the authors present a multi-target 3D RESI imaging assay for the nanomete...
www.nature.com
Reposted by JungmannLab
JungmannLab
@jungmannlab.bsky.social
· May 7
JungmannLab
@jungmannlab.bsky.social
· May 7
JungmannLab
@jungmannlab.bsky.social
· May 7