Kihara Laboratory
@kiharalab.bsky.social
120 followers
300 following
16 posts
Bioinformatics, protein modeling, cryoEM, drug screening, function prediction. Daisuke Kihara, professor of Biol/CS, Purdue U. https://kiharalab.org/ YouTube: http://alturl.com/gxvah
Posts
Media
Videos
Starter Packs
Reposted by Kihara Laboratory
Reposted by Kihara Laboratory
Bede Portz
@ysptsps.bsky.social
· Feb 7

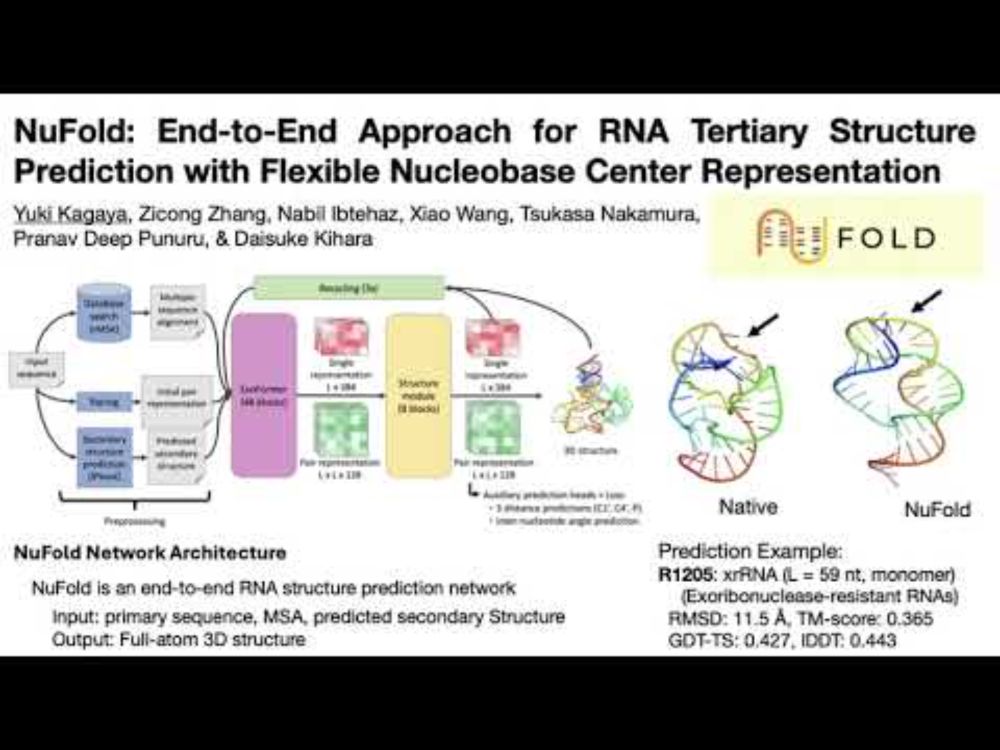
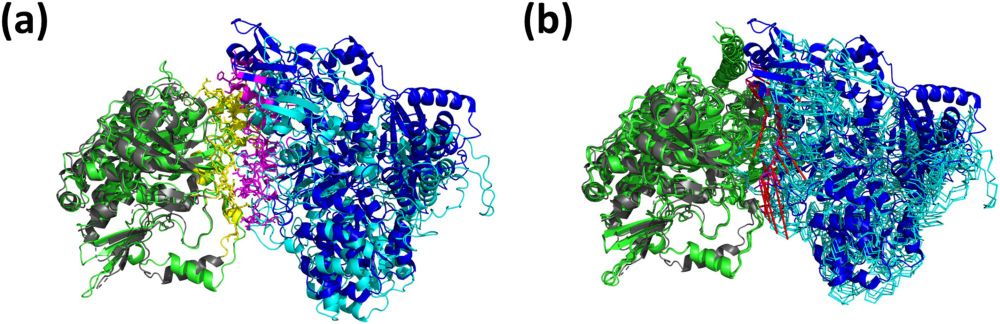
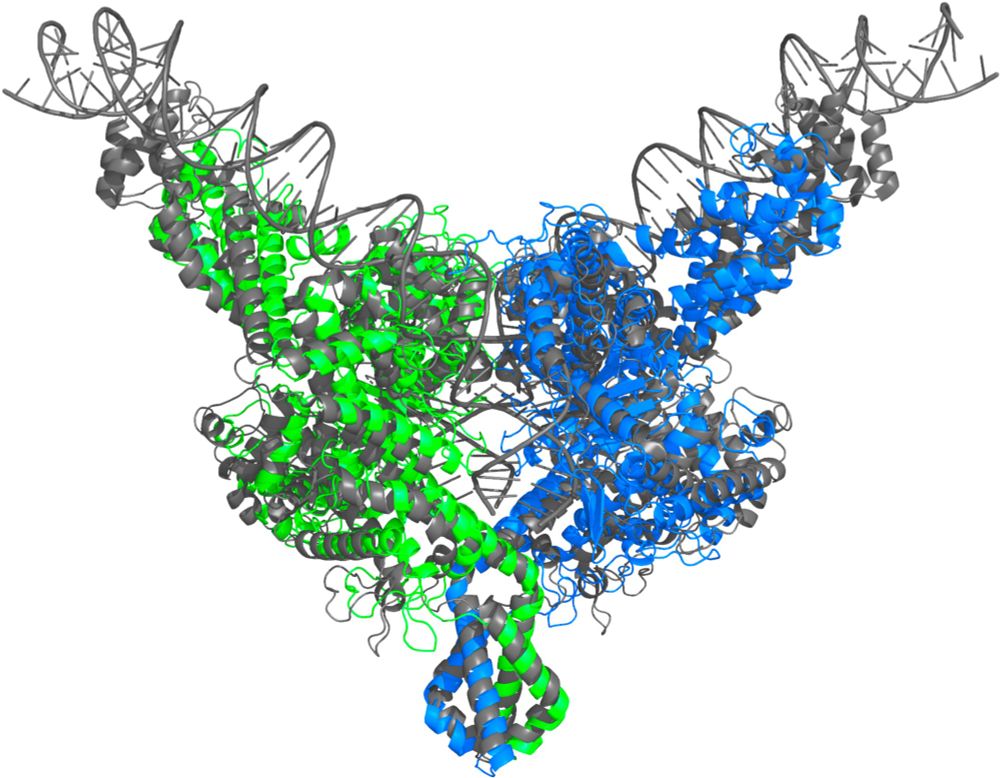
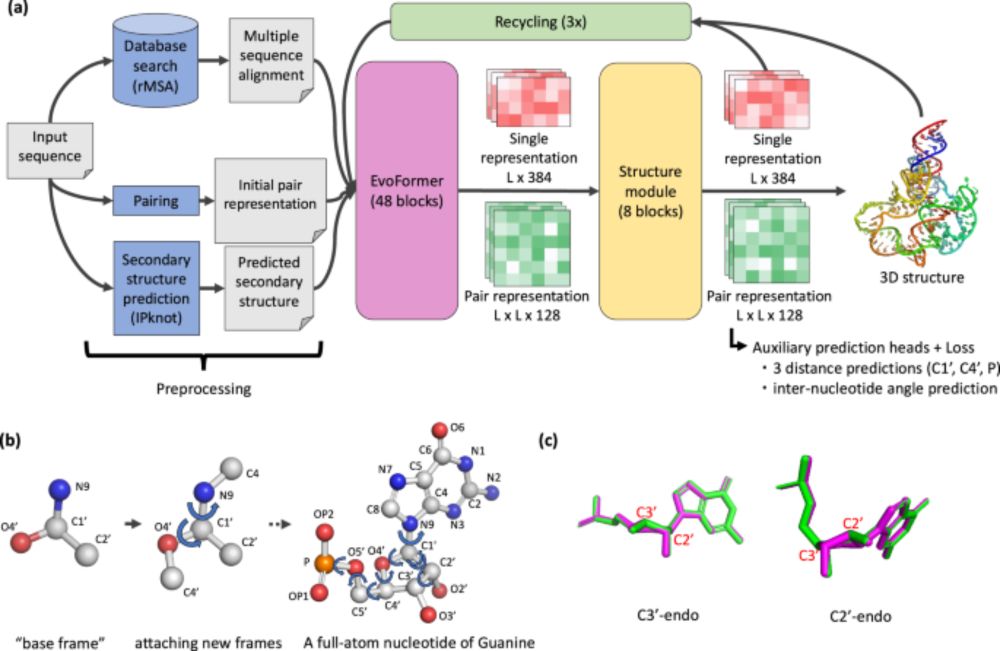
NuFold: end-to-end approach for RNA tertiary structure prediction with flexible nucleobase center representation - Nature Communications
The 3D structure of RNA is crucial for unraveling its functions. Here, the authors developed NuFold, an end-to-end deep learning-based approach that predicts all-atom RNA 3D structures from sequences....
www.nature.com
Reposted by Kihara Laboratory
Science TLDR
@sciencetldr.bsky.social
· Feb 25

NuFold: Pioneering 3D RNA Structure Prediction at Purdue University
Discover NuFold, a groundbreaking tool for predicting 3D RNA structures, developed by Purdue University. It's set to transform RNA research and expedite drug development.
www.science-tldr.com