communities.springernature.com/posts/distan...

communities.springernature.com/posts/distan...
We used DeepMainmast and DAQ for structure modeling.
pubs.acs.org/doi/full/10....

We used DeepMainmast and DAQ for structure modeling.
pubs.acs.org/doi/full/10....
daqdb.kiharalab.org
Easy to use DAQ for structure validation in your paper: em.kiharalab.org

daqdb.kiharalab.org
Easy to use DAQ for structure validation in your paper: em.kiharalab.org
www.nature.com/articles/s42...


www.nature.com/articles/s42...



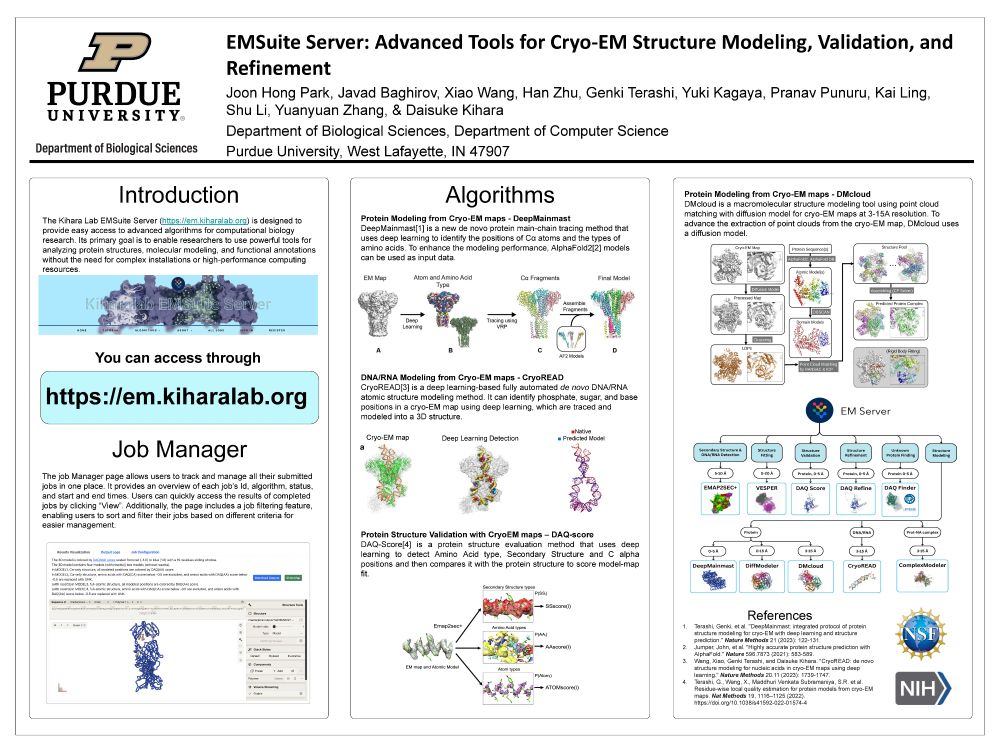
The server: em.kiharalab.org


The server: em.kiharalab.org

Visit the server for easy and accurate modeling at em.kiharalab.org
Visit the server for easy and accurate modeling at em.kiharalab.org
Paper: pubs.acs.org/doi/10.1021/...
Github: github.com/kiharalab/Di...
Google Colab: colab.research.google.com/drive/1Q1ecU...
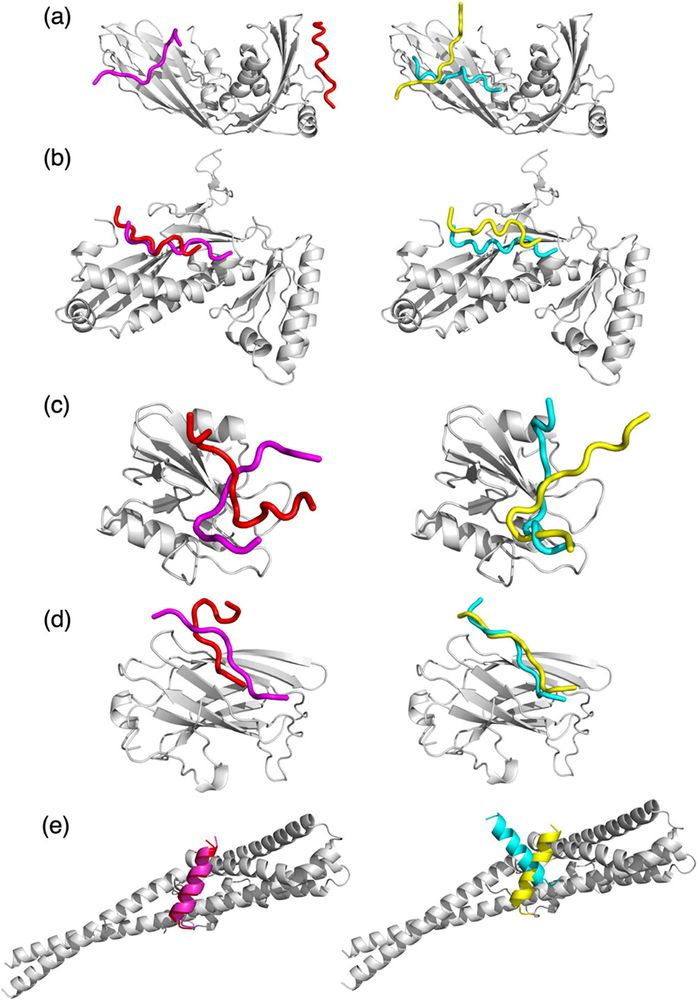
Paper: pubs.acs.org/doi/10.1021/...
Github: github.com/kiharalab/Di...
Google Colab: colab.research.google.com/drive/1Q1ecU...
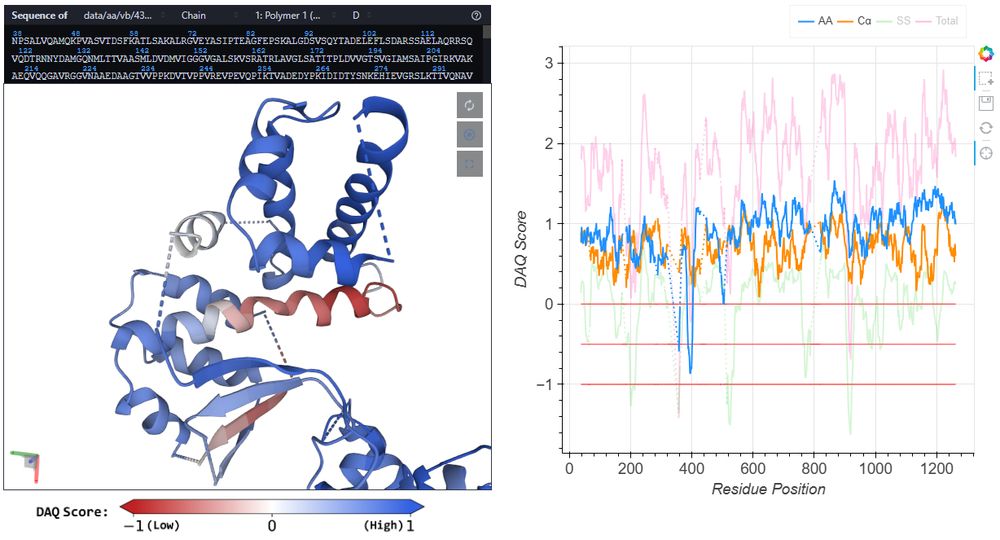
Each entry shows modeling errors in red on the structure plus chain-wise plots.
Check it out: daqdb.kiharalab.org
You can compute DAQ at: em.kiharalab.org
Each entry shows modeling errors in red on the structure plus chain-wise plots.
Check it out: daqdb.kiharalab.org
You can compute DAQ at: em.kiharalab.org
www.nsf.gov/news/using-m...
www.nsf.gov/news/using-m...
www.youtube.com/watch?v=IJOn...
You can run Nufold at Google Colab:
colab.research.google.com/github/kihar...
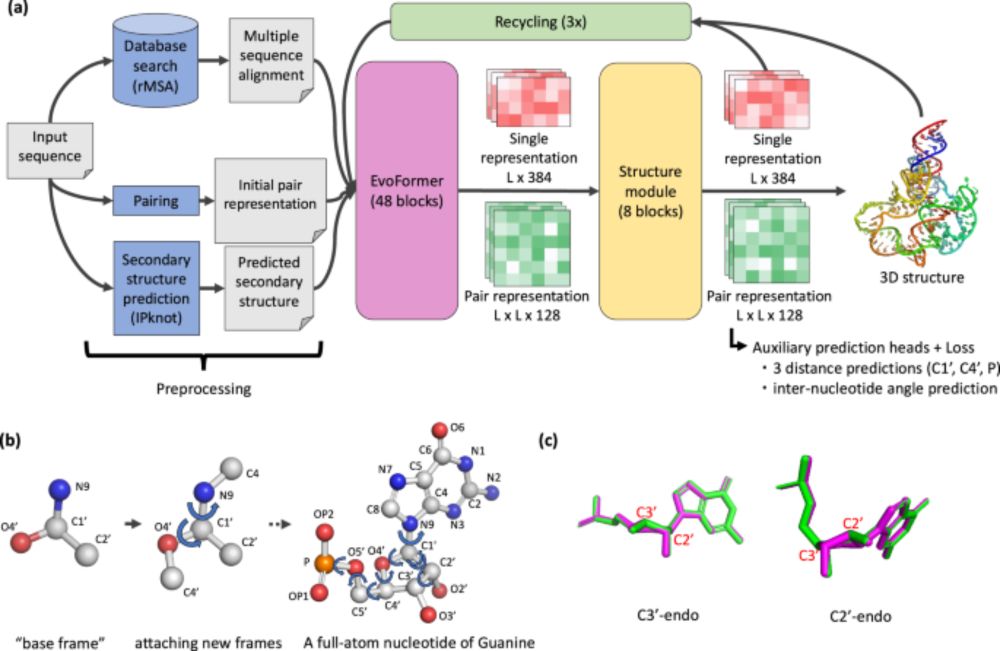
Paper: www.nature.com/articles/s41...
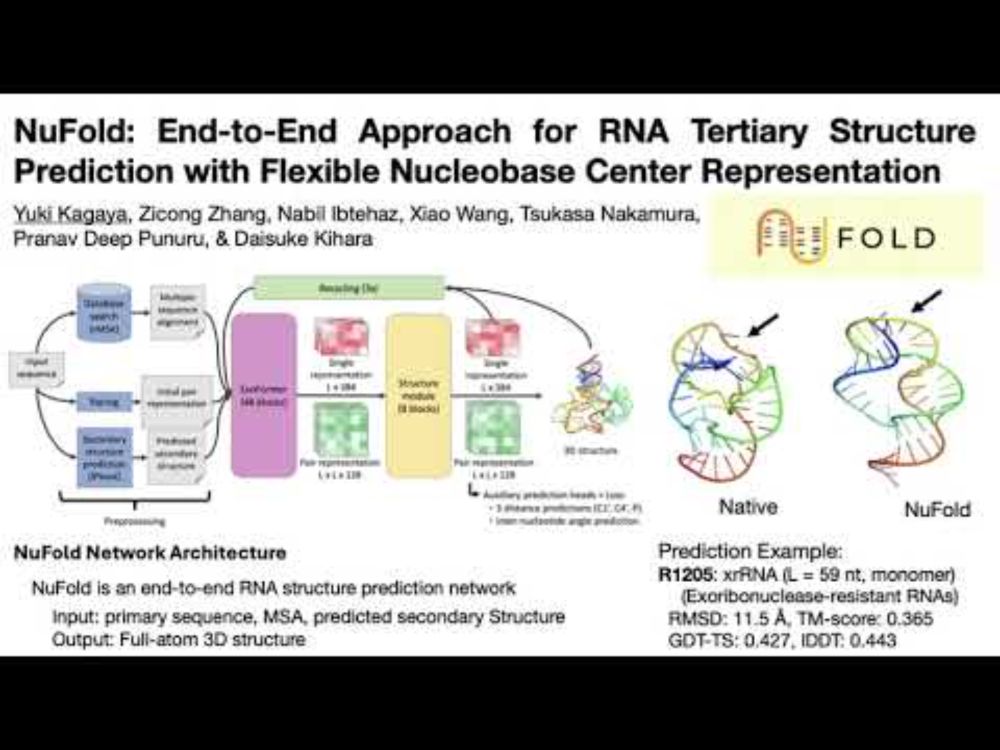
www.youtube.com/watch?v=IJOn...
You can run Nufold at Google Colab:
colab.research.google.com/github/kihar...
Paper: www.nature.com/articles/s41...
SBGrid. Here is a new tutorial video on DeepMainmast protein modeling and DAQ structure validation score presented at SBGrid Webinar:
youtu.be/z2ep8HilF-c
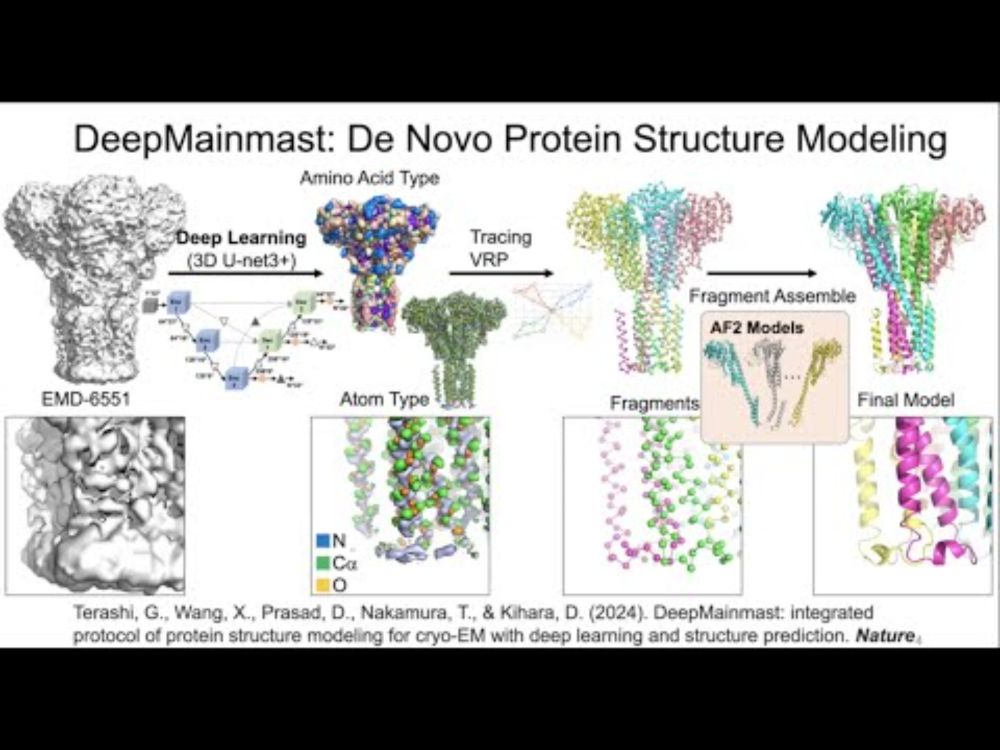
SBGrid. Here is a new tutorial video on DeepMainmast protein modeling and DAQ structure validation score presented at SBGrid Webinar:
youtu.be/z2ep8HilF-c
www.youtube.com/watch?v=YRAT...
Webserver: em.kiharalab.org/algorithm/da...

www.youtube.com/watch?v=YRAT...
Webserver: em.kiharalab.org/algorithm/da...
onlinelibrary.wiley.com/doi/10.1002/...
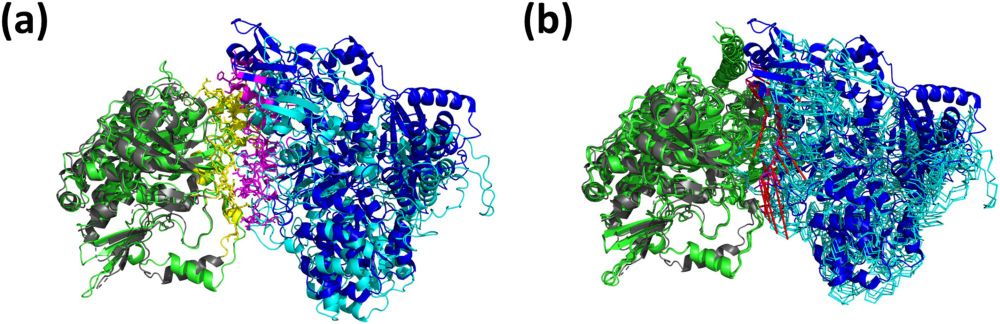
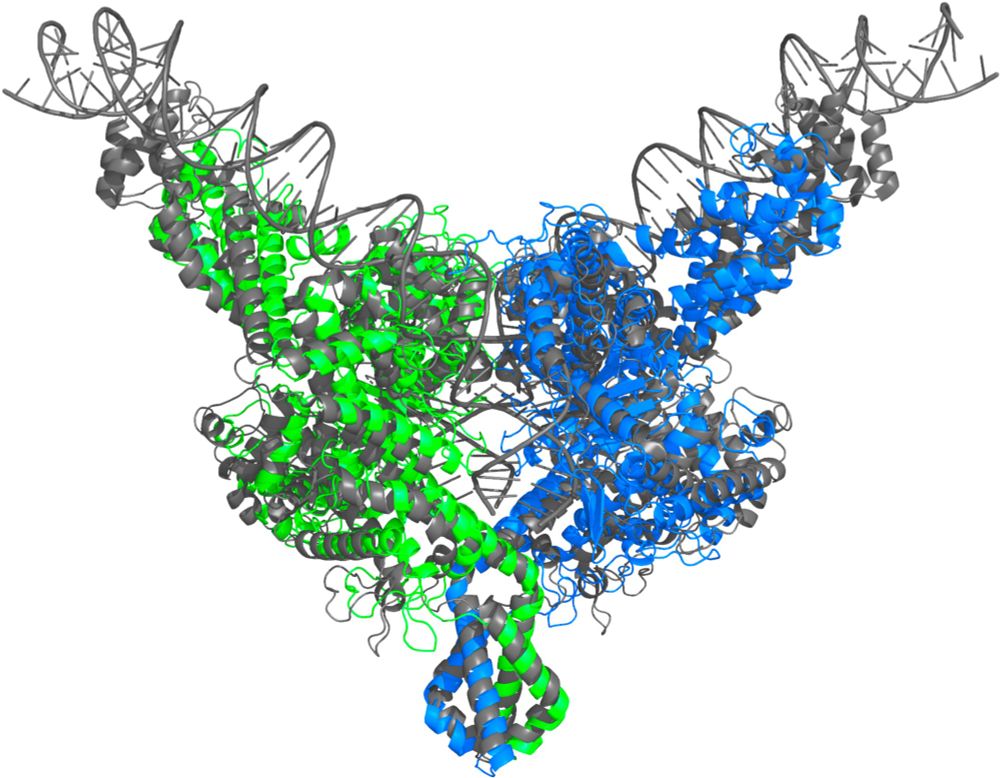
onlinelibrary.wiley.com/doi/10.1002/...
If you missed the initial deadline, this is your chance to present your research and engage with the bioinformatics and computational biology community.
📥Submit now: https://t.ly/ByJrY

If you missed the initial deadline, this is your chance to present your research and engage with the bioinformatics and computational biology community.
📥Submit now: https://t.ly/ByJrY