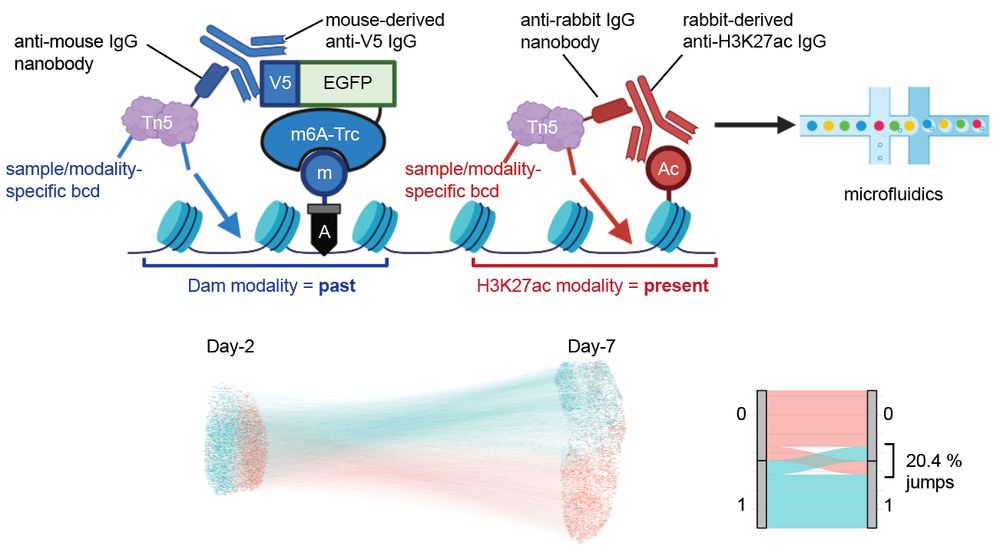
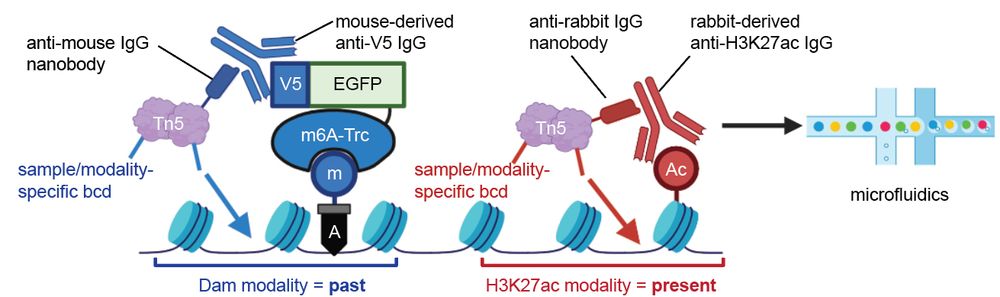
Taro Kitazawa
@kitazawa-taro.bsky.social
150 followers
160 following
27 posts
Group Leader at DANDRITE Nordic-EMBL/Aarhus Univ.
ERC Research, FensKavli Scholar.
Molecular basis of neuroplasticity.
New genomics technologies.
https://www.kitazawa-lab.com/home
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Taro Kitazawa
Taro Kitazawa
@kitazawa-taro.bsky.social
· Aug 18
Taro Kitazawa
@kitazawa-taro.bsky.social
· Aug 16
Taro Kitazawa
@kitazawa-taro.bsky.social
· Aug 16
Reposted by Taro Kitazawa
Taro Kitazawa
@kitazawa-taro.bsky.social
· Aug 16
Taro Kitazawa
@kitazawa-taro.bsky.social
· Aug 16
Taro Kitazawa
@kitazawa-taro.bsky.social
· Aug 16
Taro Kitazawa
@kitazawa-taro.bsky.social
· Aug 16
Taro Kitazawa
@kitazawa-taro.bsky.social
· Jul 24