Konopka Lab
@konopkalab.bsky.social
110 followers
120 following
16 posts
Official Konopka Lab Account!
We're a group of spirited scientists exploring the genetic landscape of the evolved human brain.
@Los Angeles, CA, konopkalab.org
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Konopka Lab
Waggoner Lab
@labwaggoner.bsky.social
· Jun 30

Human CLOCK enhances neocortical function - Nature Neuroscience
Regulation of gene expression is a facet of human brain specialization. Here, the authors show that human-like expression of the CLOCK gene in the mouse neocortex enhances cognitive flexibility and ne...
www.nature.com
Konopka Lab
@konopkalab.bsky.social
· Apr 23
Konopka Lab
@konopkalab.bsky.social
· Apr 22
Konopka Lab
@konopkalab.bsky.social
· Apr 22
Konopka Lab
@konopkalab.bsky.social
· Apr 22
Konopka Lab
@konopkalab.bsky.social
· Apr 22
Konopka Lab
@konopkalab.bsky.social
· Apr 22
Konopka Lab
@konopkalab.bsky.social
· Apr 22
Konopka Lab
@konopkalab.bsky.social
· Apr 22
Konopka Lab
@konopkalab.bsky.social
· Apr 22
Konopka Lab
@konopkalab.bsky.social
· Apr 22
Konopka Lab
@konopkalab.bsky.social
· Apr 22

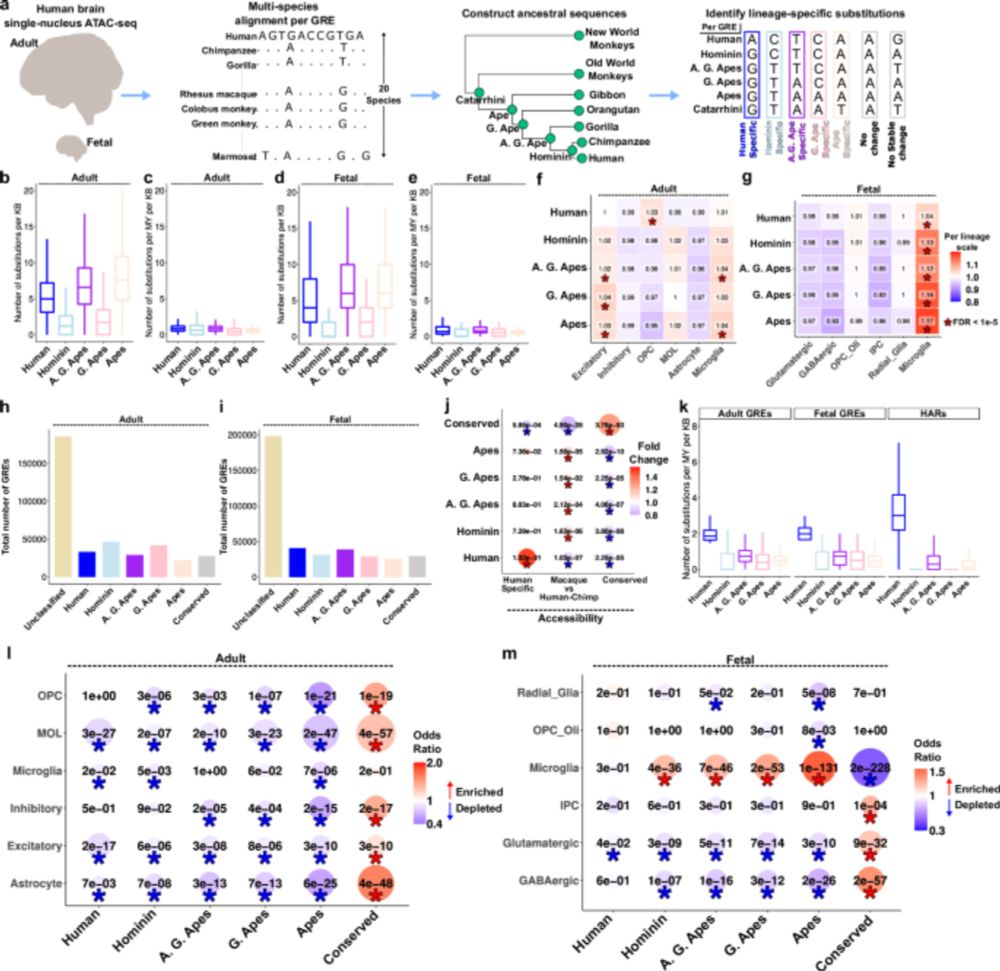
Evolution of the cellular landscape in mammalian striatum
The dorsal striatum is important for highly specialized functions including movement, learning, and habit formation. However, it is not known if species-specialized behaviors are associated with cellu...
www.biorxiv.org
Konopka Lab
@konopkalab.bsky.social
· Apr 22
Reposted by Konopka Lab
Cedric Boeckx
@cedricboeckx.bsky.social
· Apr 21

Evolution of the cellular landscape in mammalian striatum
The dorsal striatum is important for highly specialized functions including movement, learning, and habit formation. However, it is not known if species-specialized behaviors are associated with cellu...
www.biorxiv.org