Konrad Chudzik
@konrad-chudzik.bsky.social
190 followers
310 following
16 posts
PhD Candidate in Regulatory Genome program (IRTG2403) |
Robson lab - Berlin Institute for Medical Systems Biology (MDC-BIMSB) & Mundlos Lab - Max Planck Institute for Molecular Genetics |
Genome Regulation, Nuclear Envelope, LADs
Posts
Media
Videos
Starter Packs
Reposted by Konrad Chudzik
Reposted by Konrad Chudzik
Abby Buchwalter
@abbybuch.bsky.social
· Jul 22
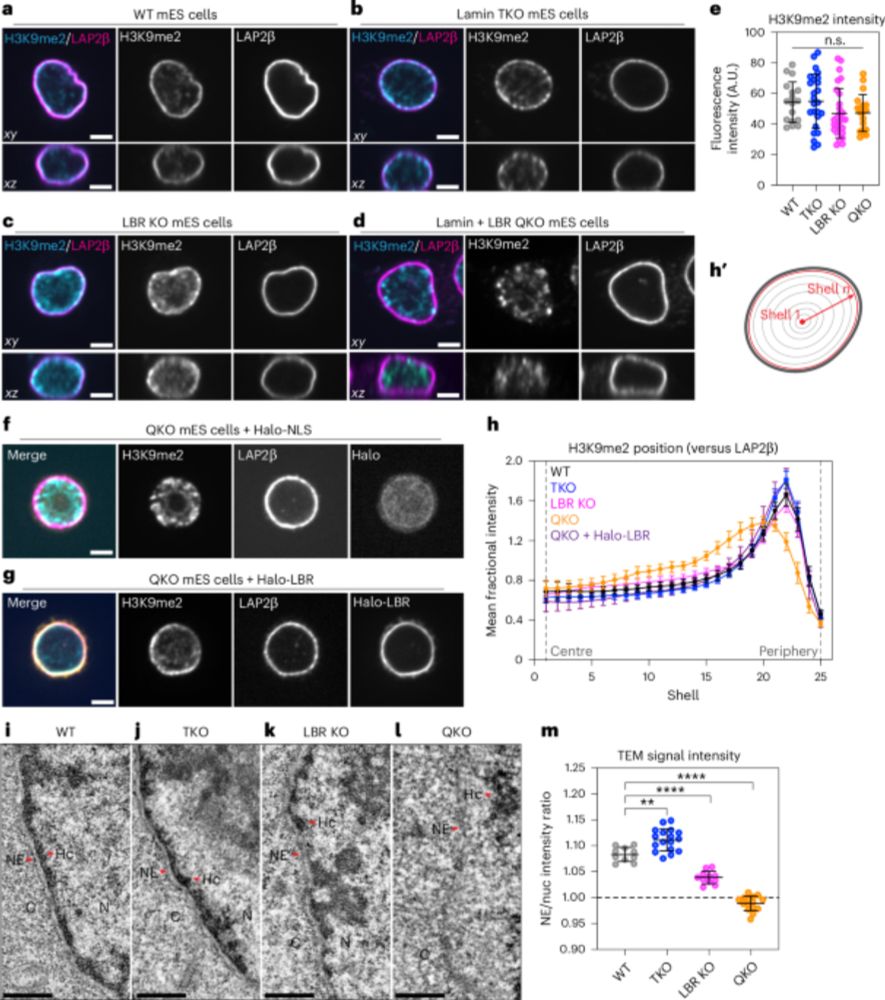
The nuclear periphery confers repression on H3K9me2-marked genes and transposons to shape cell fate - Nature Cell Biology
Marin et al. report the role of lamin proteins and the lamin B receptor (LBR) in chromatin positioning at the nuclear periphery. Knockout of all lamins and LBR in mouse embryonic stem cells leads to h...
www.nature.com
Reposted by Konrad Chudzik
Jop Kind
@jopkind.bsky.social
· Jul 10
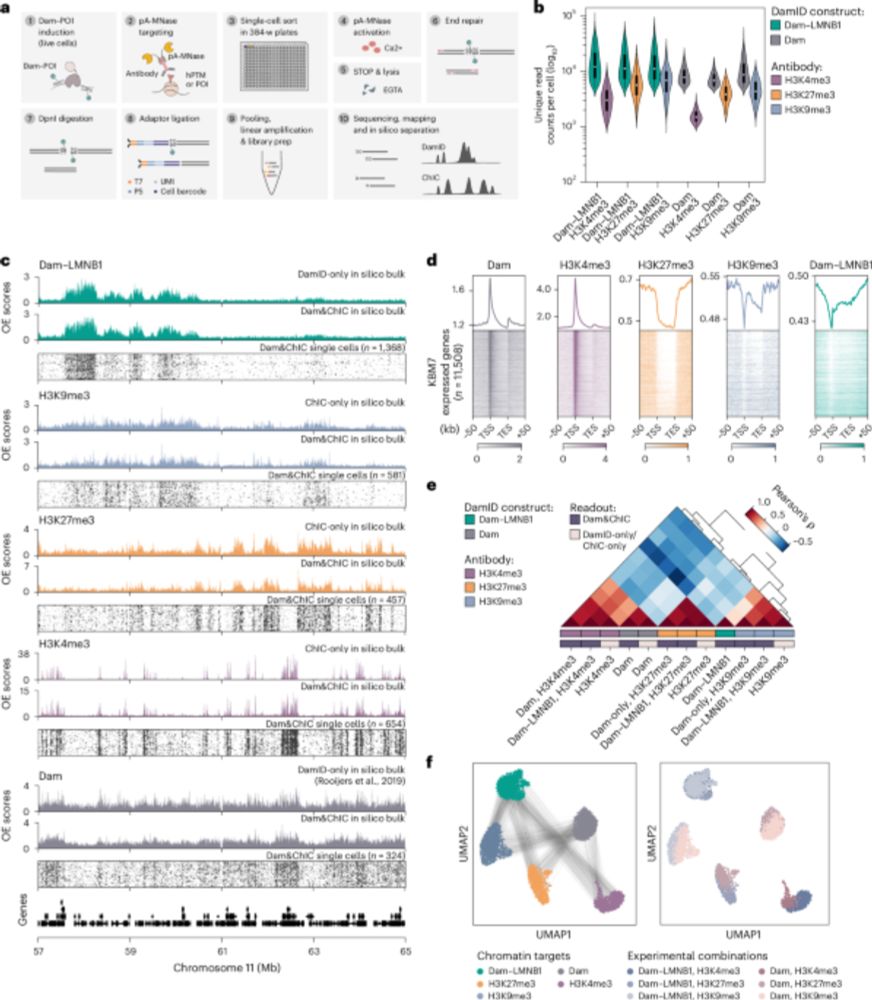
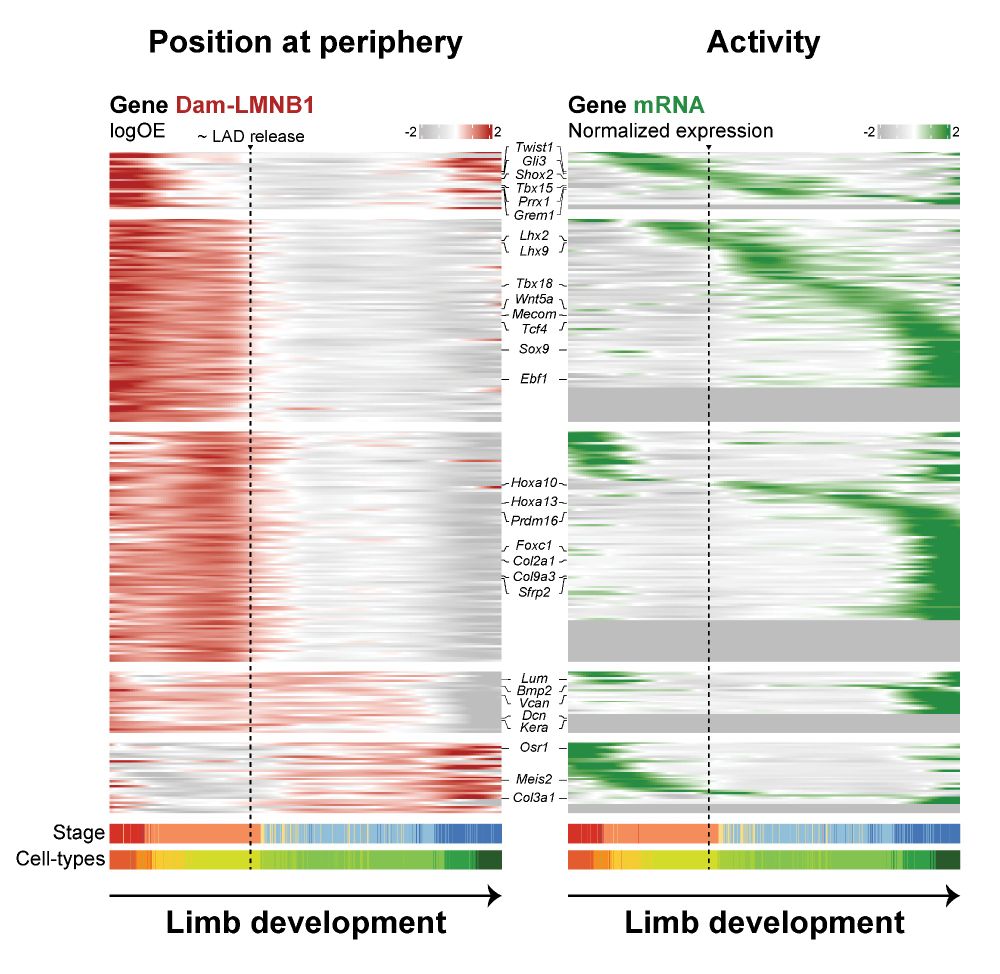
Retrospective and multifactorial single-cell profiling reveals sequential chromatin reorganization during X inactivation - Nature Cell Biology
Kefalopoulou, Rullens et al. develop Dam&ChIC to assay chromatin state at two different time points in the same cell. The method was used to study the reorganization of LADs during cell division a...
www.nature.com
Reposted by Konrad Chudzik
Juliane Glaser
@julianeg.bsky.social
· Jul 9
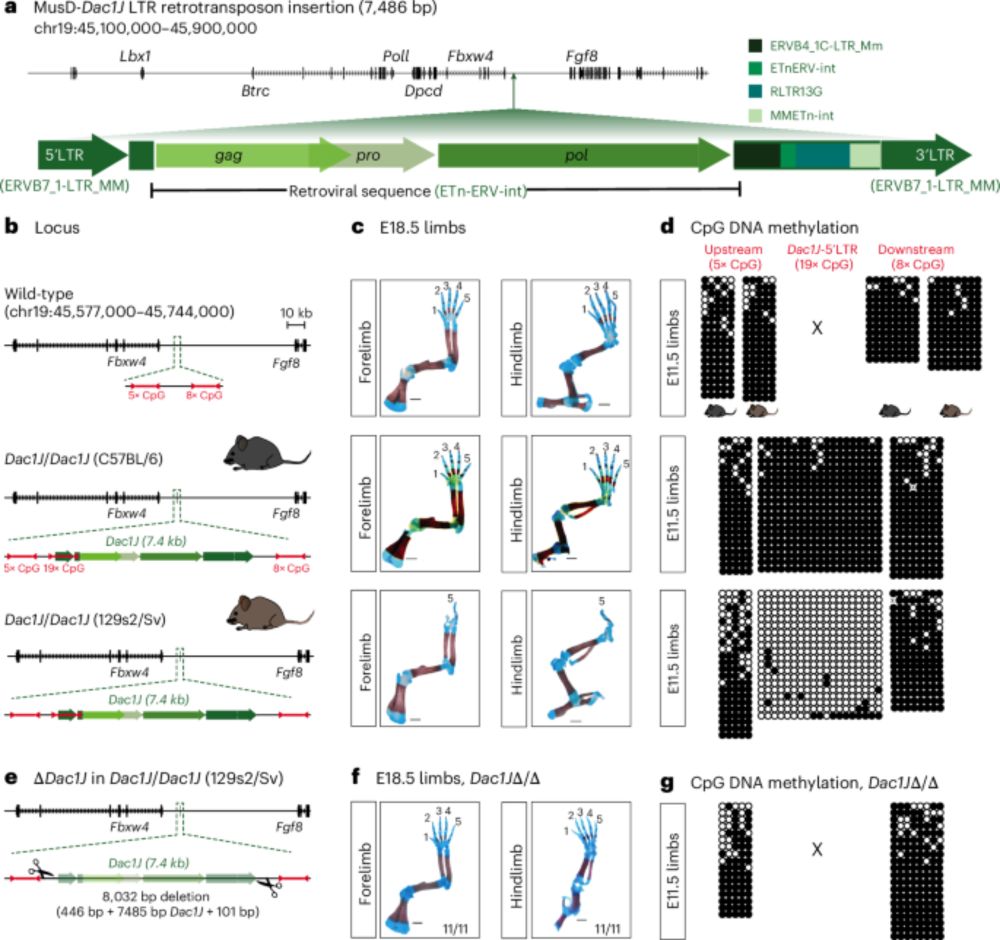
Enhancer adoption by an LTR retrotransposon generates viral-like particles, causing developmental limb phenotypes - Nature Genetics
Activation of an LTR retrotransposon inserted upstream of the Fgf8 gene produces viral-like particles in the mouse developing limb, triggering apoptosis and causing limb malformation. This phenotype c...
www.nature.com
Reposted by Konrad Chudzik
Mike Robson
@drmrobson.bsky.social
· Jul 8
Reposted by Konrad Chudzik
Reposted by Konrad Chudzik
Daniel Ibrahim
@danielibrahim.bsky.social
· May 27

Conservation of regulatory elements with highly diverged sequences across large evolutionary distances
Nature Genetics - Combining functional genomic data from mouse and chicken with a synteny-based strategy identifies positionally conserved cis-regulatory elements in the absence of direct sequence...
rdcu.be
Reposted by Konrad Chudzik
Mike Robson
@drmrobson.bsky.social
· May 31
Reposted by Konrad Chudzik
Reposted by Konrad Chudzik
Iana V. Kim
@ianakim.bsky.social
· May 7
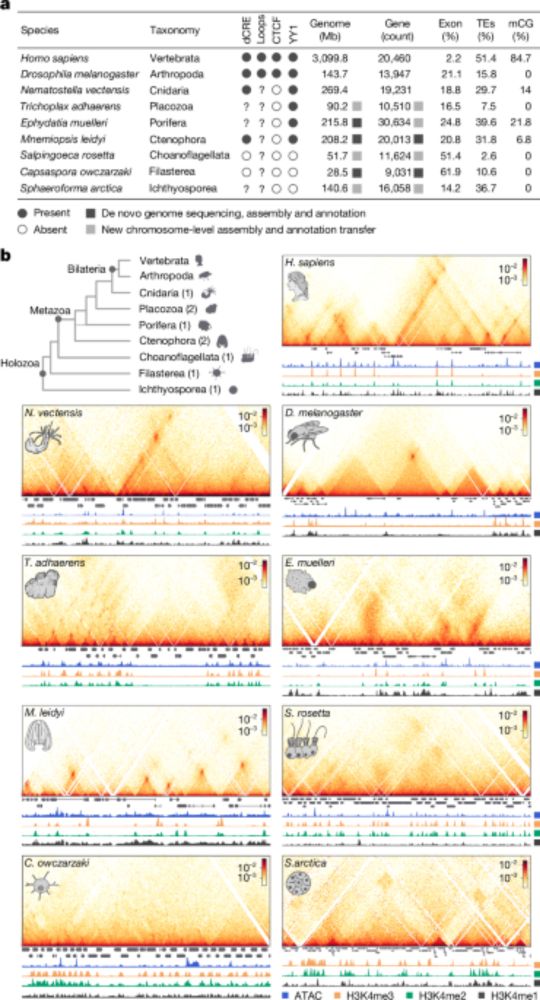
Chromatin loops are an ancestral hallmark of the animal regulatory genome - Nature
The physical organization of the genome in non-bilaterian animals and their closest unicellular relatives is characterized; comparative analysis shows chromatin looping is a conserved feature of ...
www.nature.com
Reposted by Konrad Chudzik