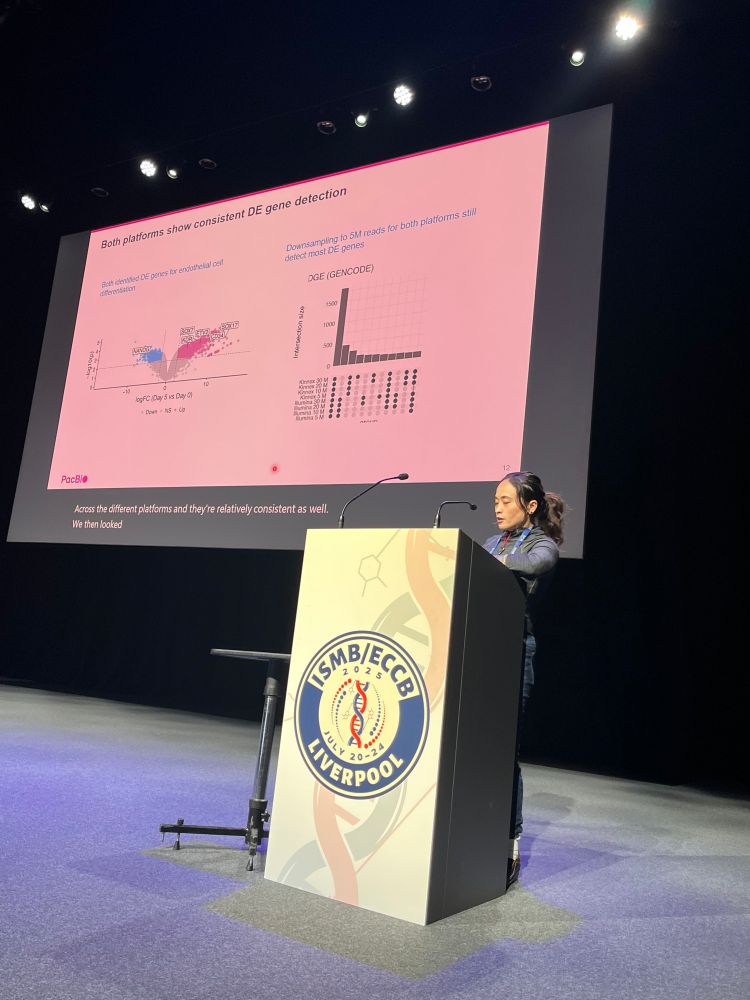
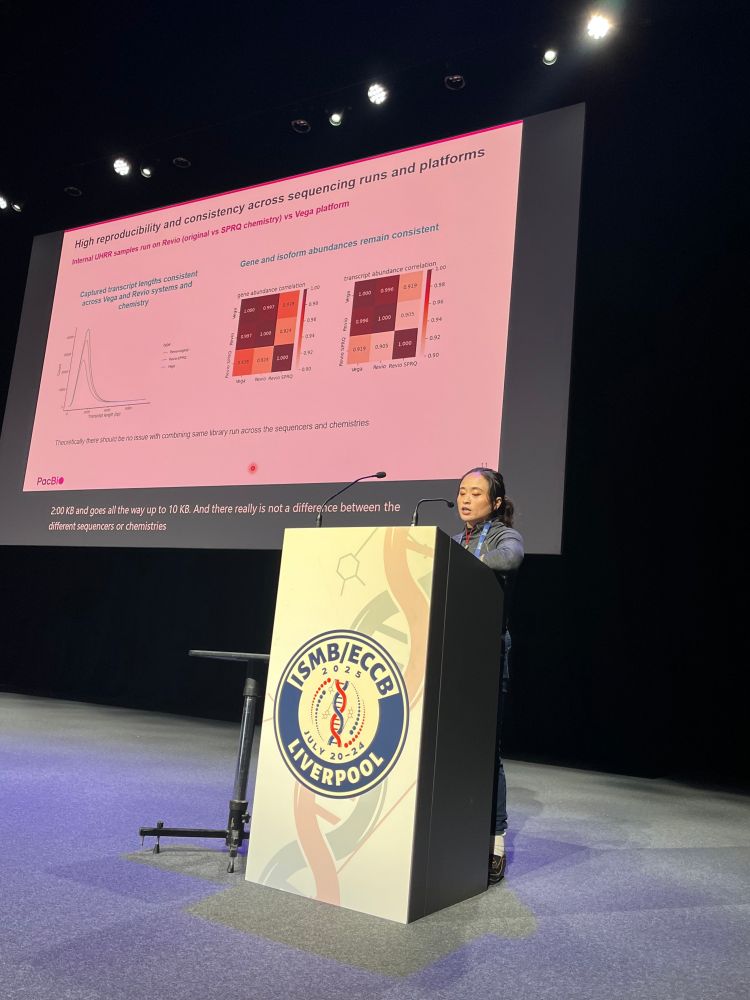
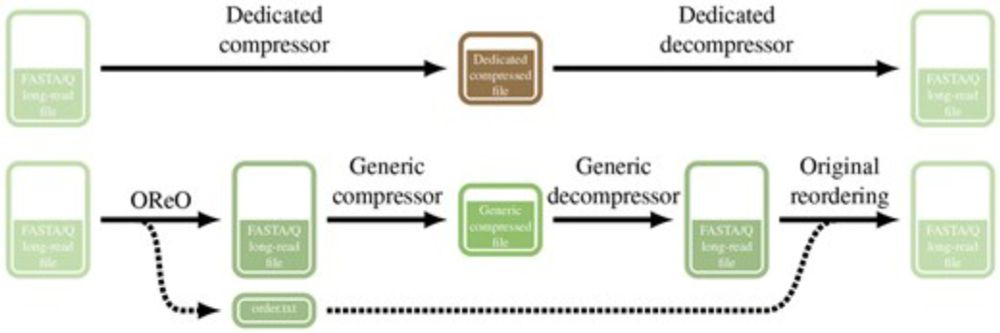
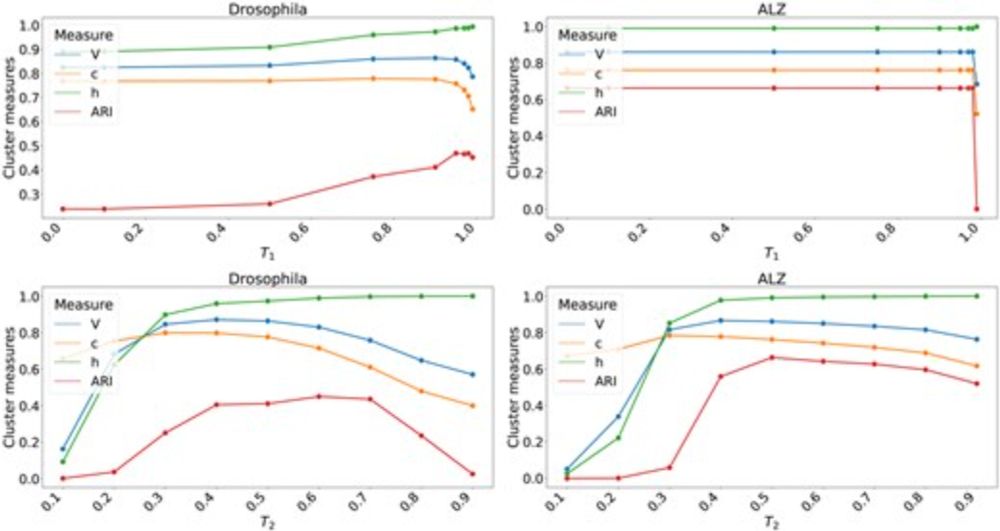
Kristoffer Sahlin
@ksahlin.bsky.social
750 followers
150 following
23 posts
Assistant Professor at the Department of Mathematics, Stockholm University, and a Scilifelab Fellow. Algorithms, Modeling, Transcriptomics, Genomics.
Amateur runner 5000m 18:48 | 10k 37:40 | HM 1:28:34 | M 3:39:06
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Kristoffer Sahlin
Reposted by Kristoffer Sahlin
Reposted by Kristoffer Sahlin
Kristoffer Sahlin
@ksahlin.bsky.social
· Jul 25
Reposted by Kristoffer Sahlin
Reposted by Kristoffer Sahlin
Reposted by Kristoffer Sahlin
Reposted by Kristoffer Sahlin
Reposted by Kristoffer Sahlin
Reposted by Kristoffer Sahlin
Reposted by Kristoffer Sahlin
Reposted by Kristoffer Sahlin
Paul Medvedev
@pashadag.bsky.social
· Jun 25
Reposted by Kristoffer Sahlin
Reposted by Kristoffer Sahlin
Kristoffer Sahlin
@ksahlin.bsky.social
· Apr 25
Reposted by Kristoffer Sahlin