Kaitlin Samocha
@ksamocha.bsky.social
1.5K followers
230 following
58 posts
Assistant Investigator @ MGH / Broad / HMS. Focus on human genomics and modeling rare variation. She/her
Posts
Media
Videos
Starter Packs
Reposted by Kaitlin Samocha
Reposted by Kaitlin Samocha
Reposted by Kaitlin Samocha
Reposted by Kaitlin Samocha
Alex Hoischen
@ahoischen.bsky.social
· May 24
Reposted by Kaitlin Samocha
Zornitza Stark
@zornitza.bsky.social
· May 23

Scalable automated reanalysis of genomic data in research and clinical rare disease cohorts
Reanalysis of genomic data in rare disease is highly effective in increasing diagnostic yields but remains limited by manual approaches. Automation and optimization for high specificity will be necess...
www.medrxiv.org
Reposted by Kaitlin Samocha
Kaitlin Samocha
@ksamocha.bsky.social
· Feb 21
Reposted by Kaitlin Samocha
Kaitlin Samocha
@ksamocha.bsky.social
· Apr 19
Kaitlin Samocha
@ksamocha.bsky.social
· Apr 19
Kaitlin Samocha
@ksamocha.bsky.social
· Mar 8
Kaitlin Samocha
@ksamocha.bsky.social
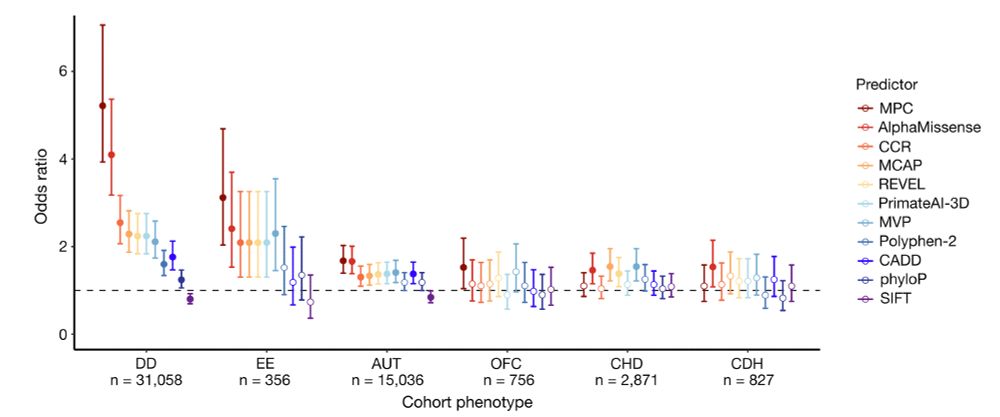
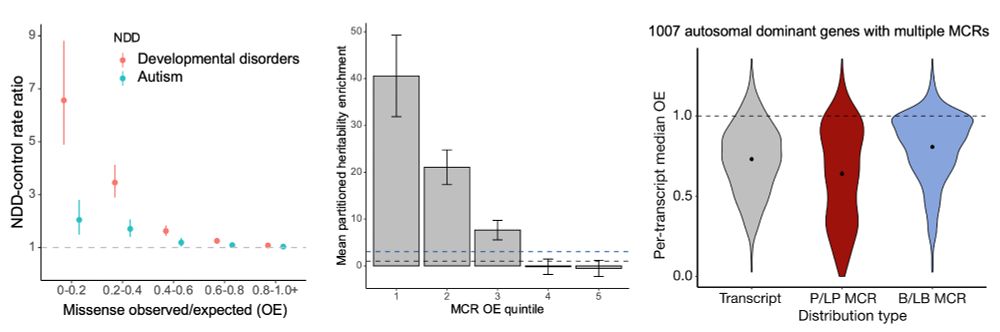
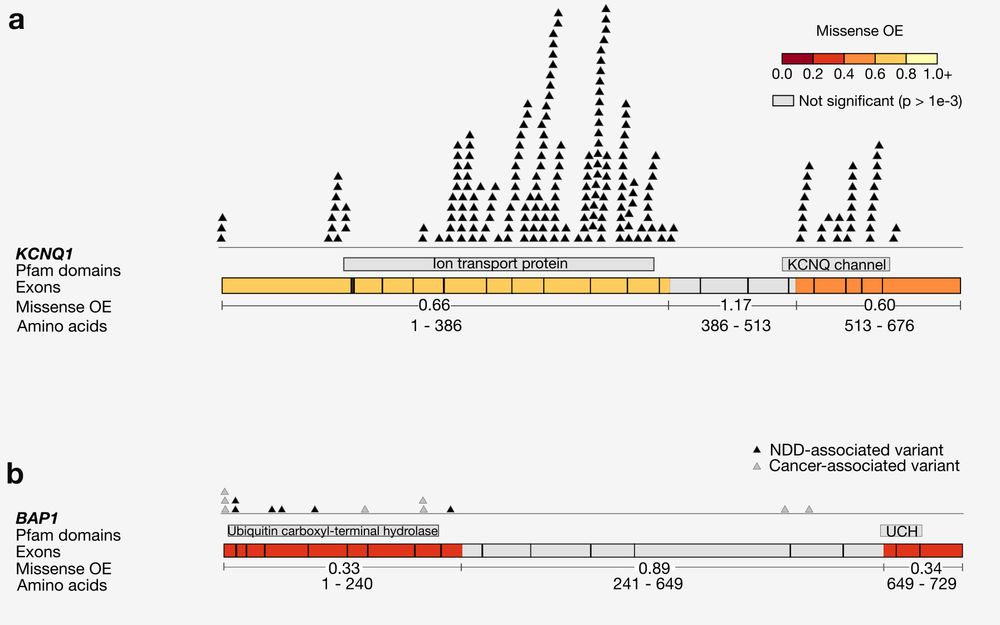
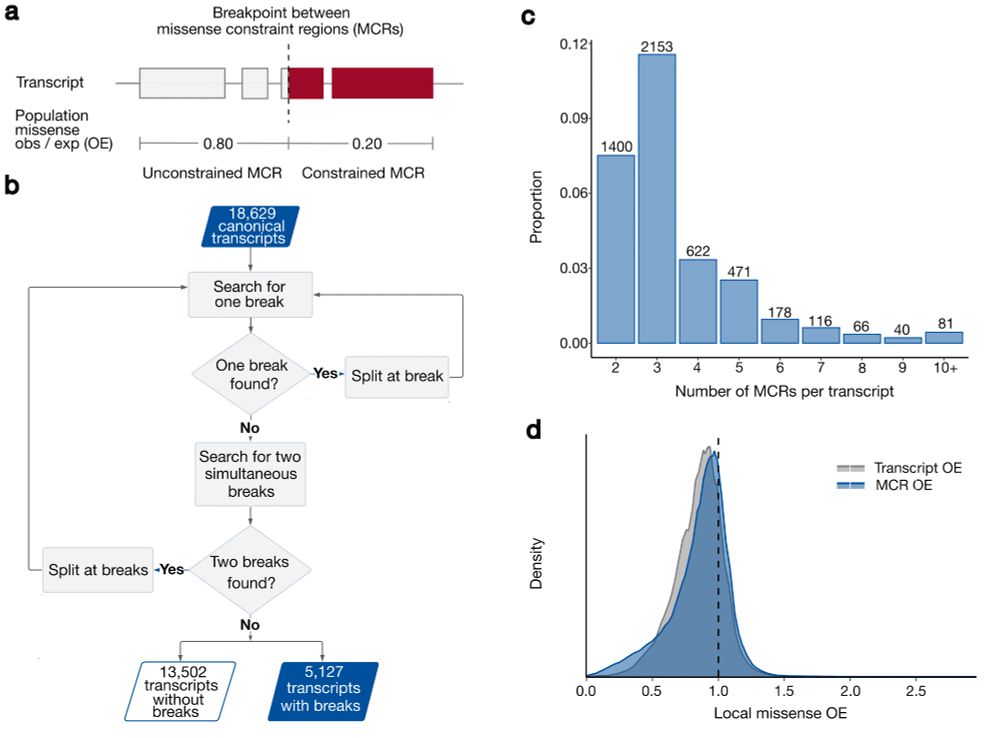
· Dec 8
Kaitlin Samocha
@ksamocha.bsky.social
· Dec 8