"More thinking and less pipetting"




www.biorxiv.org/content/10.6...
www.biorxiv.org/content/10.6...

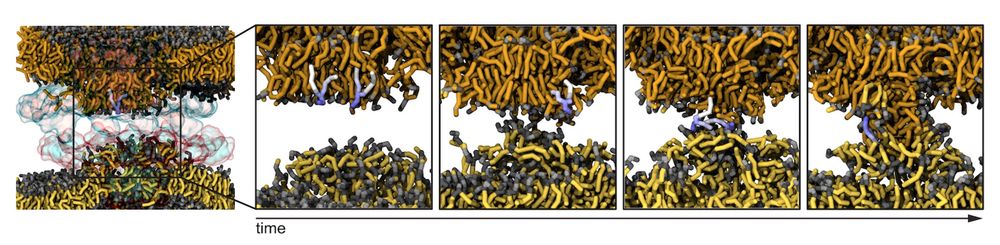
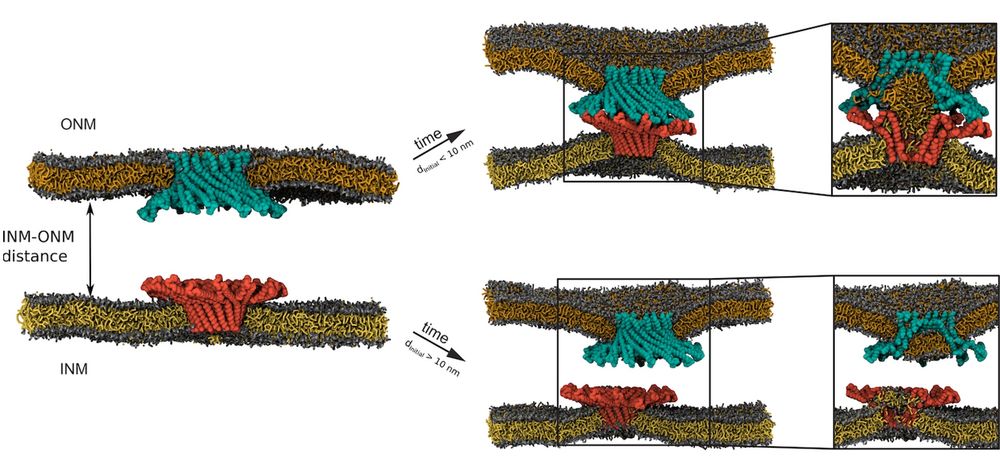
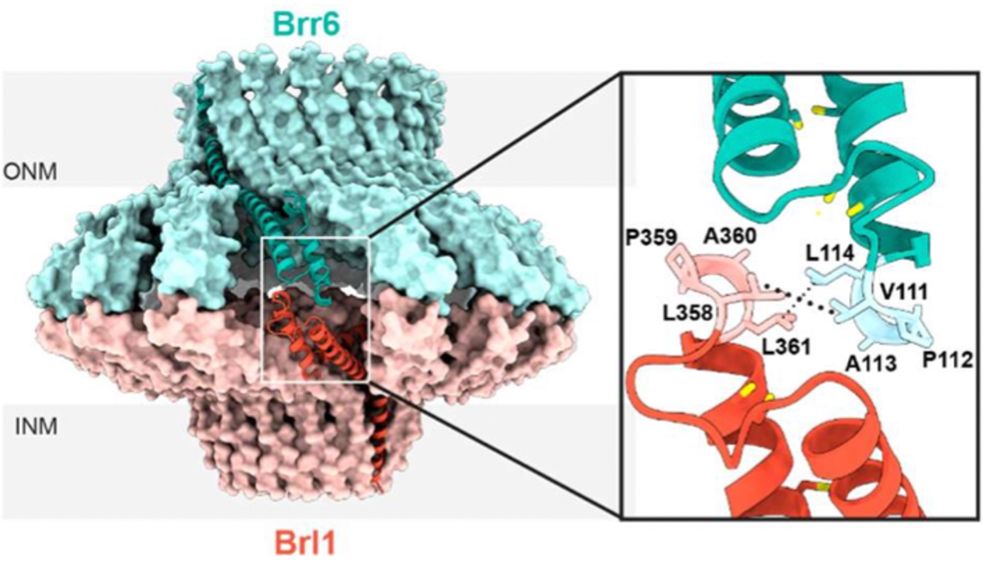
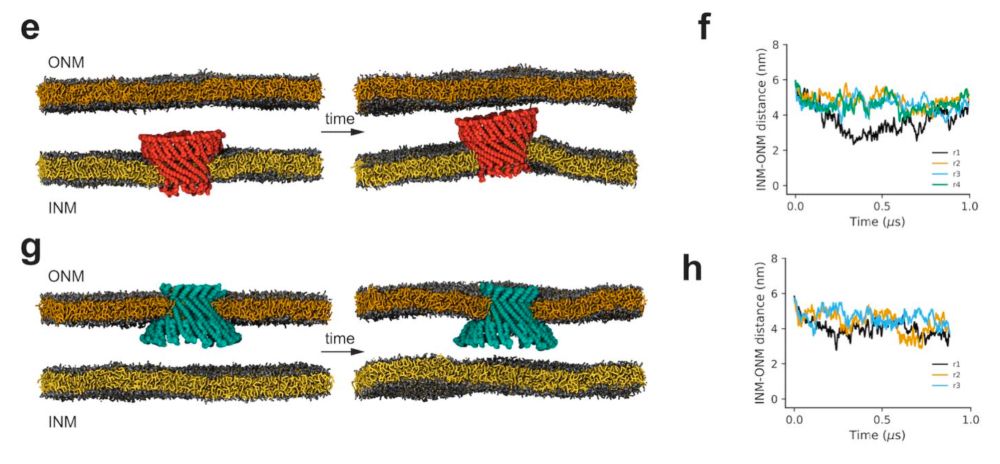
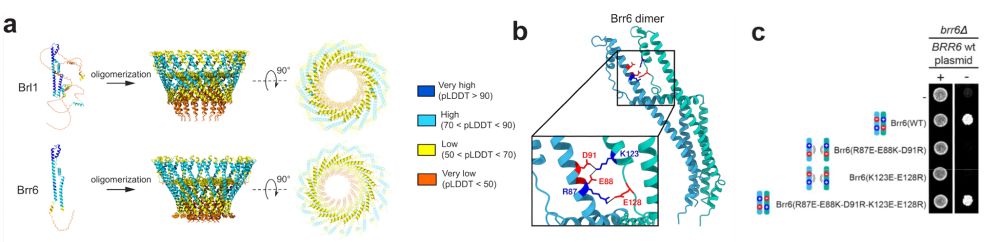
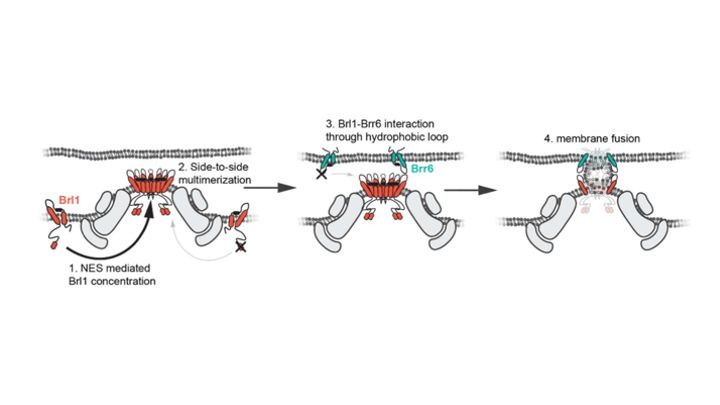
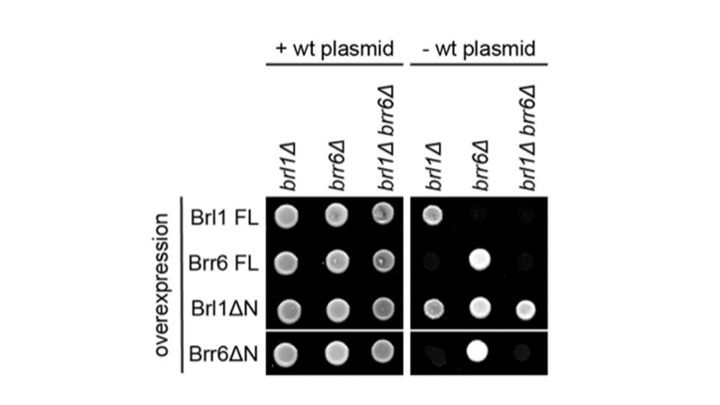
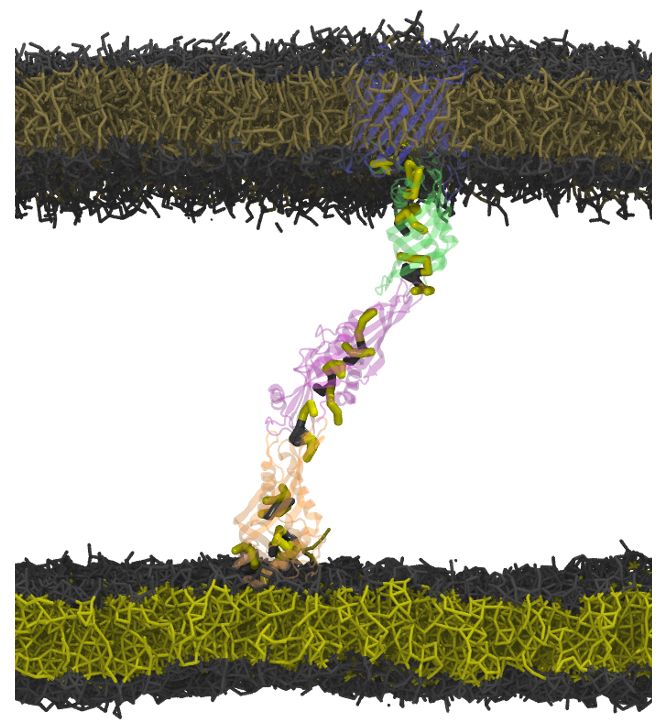
How do nuclear membranes fuse during NPC assembly? We answer this question in our latest work, where we identify a new mechanism for membrane fusion… (1/13)
How do nuclear membranes fuse during NPC assembly? We answer this question in our latest work, where we identify a new mechanism for membrane fusion… (1/13)